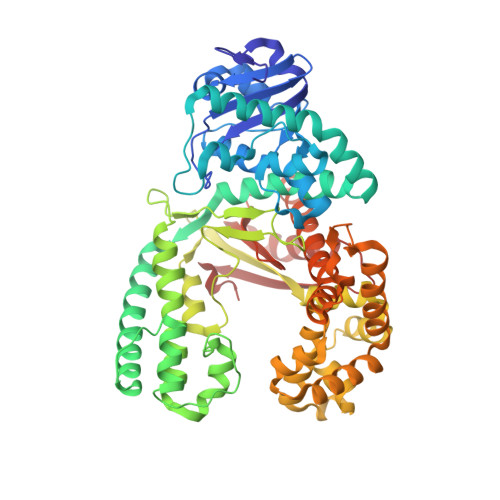
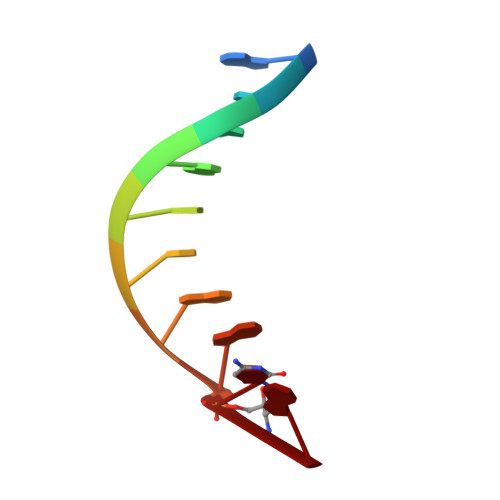
Synthesis of phosphoramidate-linked DNA by a modified DNA polymerase.
Lelyveld, V.S., Zhang, W., Szostak, J.W.(2020) Proc Natl Acad Sci U S A 117: 7276-7283
- PubMed: 32188786
- DOI: https://doi.org/10.1073/pnas.1922400117
- Primary Citation of Related Structures:
6UR2, 6UR4, 6UR9, 6US5 - PubMed Abstract:
All known polymerases copy genetic material by catalyzing phosphodiester bond formation. This highly conserved activity proceeds by a common mechanism, such that incorporated nucleoside analogs terminate chain elongation if the resulting primer strand lacks a terminal hydroxyl group. Even conservatively substituted 3'-amino nucleotides generally act as chain terminators, and no enzymatic pathway for their polymerization has yet been found. Although 3'-amino nucleotides can be chemically coupled to yield stable oligonucleotides containing N3'→P5' phosphoramidate (NP) bonds, no such internucleotide linkages are known to occur in nature. Here, we report that 3'-amino terminated primers are, in fact, slowly extended by the DNA polymerase from B. stearothermophilus in a template-directed manner. When its cofactor is Ca 2+ rather than Mg 2+ , the reaction is fivefold faster, permitting multiple turnover NP bond formation to yield NP-DNA strands from the corresponding 3'-amino-2',3'-dideoxynucleoside 5'-triphosphates. A single active site mutation further enhances the rate of NP-DNA synthesis by an additional 21-fold. We show that DNA-dependent NP-DNA polymerase activity depends on conserved active site residues and propose a likely mechanism for this activity based on a series of crystal structures of bound complexes. Our results significantly broaden the catalytic scope of polymerase activity and suggest the feasibility of a genetic transition between native nucleic acids and NP-DNA.
- Department of Molecular Biology, Center for Computational and Integrative Biology, Massachusetts General Hospital, Boston, MA 02114.
Organizational Affiliation: