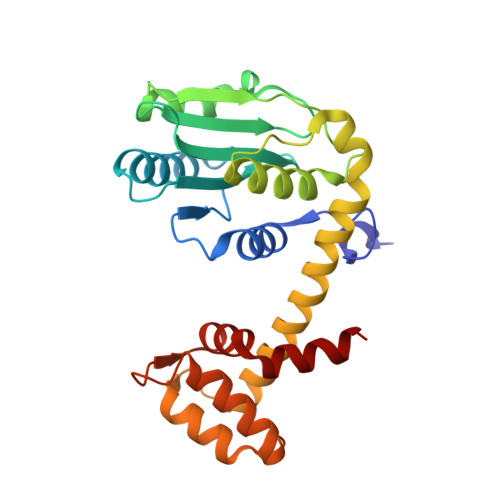
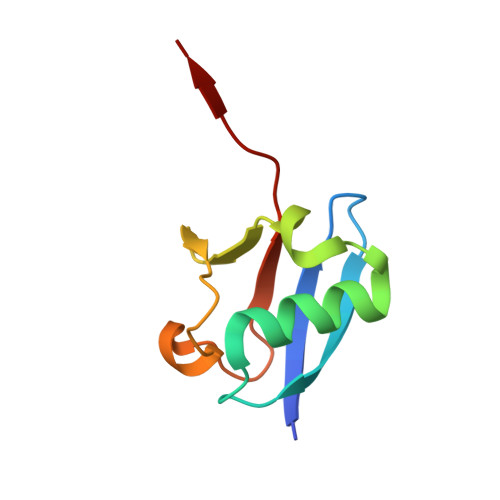
A deubiquitylase with an unusually high-affinity ubiquitin-binding domain from the scrub typhus pathogen Orientia tsutsugamushi.
Berk, J.M., Lim, C., Ronau, J.A., Chaudhuri, A., Chen, H., Beckmann, J.F., Loria, J.P., Xiong, Y., Hochstrasser, M.(2020) Nat Commun 11: 2343-2343
- PubMed: 32393759
- DOI: https://doi.org/10.1038/s41467-020-15985-4
- Primary Citation of Related Structures:
6UPS, 6UPU - PubMed Abstract:
Ubiquitin mediated signaling contributes critically to host cell defenses during pathogen infection. Many pathogens manipulate the ubiquitin system to evade these defenses. Here we characterize a likely effector protein bearing a deubiquitylase (DUB) domain from the obligate intracellular bacterium Orientia tsutsugamushi, the causative agent of scrub typhus. The Ulp1-like DUB prefers ubiquitin substrates over ubiquitin-like proteins and efficiently cleaves polyubiquitin chains of three or more ubiquitins. The co-crystal structure of the DUB (OtDUB) domain with ubiquitin revealed three bound ubiquitins: one engages the S1 site, the second binds an S2 site contributing to chain specificity and the third binds a unique ubiquitin-binding domain (UBD). The UBD modulates OtDUB activity, undergoes a pronounced structural transition upon binding ubiquitin, and binds monoubiquitin with an unprecedented ~5 nM dissociation constant. The characterization and high-resolution structure determination of this enzyme should aid in its development as a drug target to counter Orientia infections.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT, 06520, USA.
Organizational Affiliation: