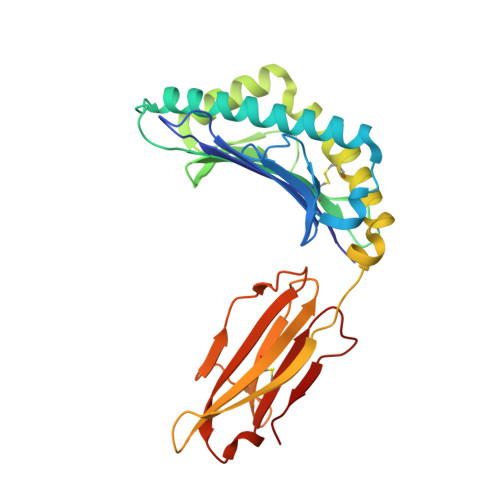
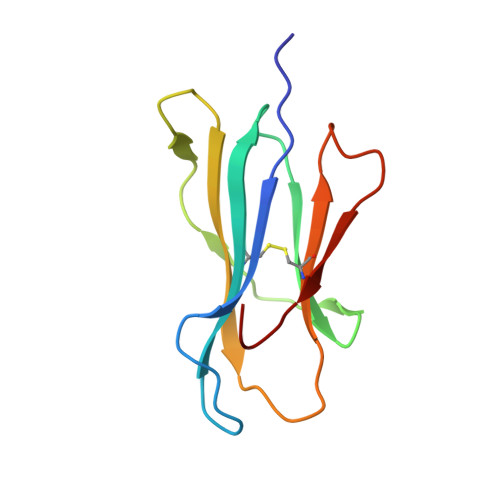
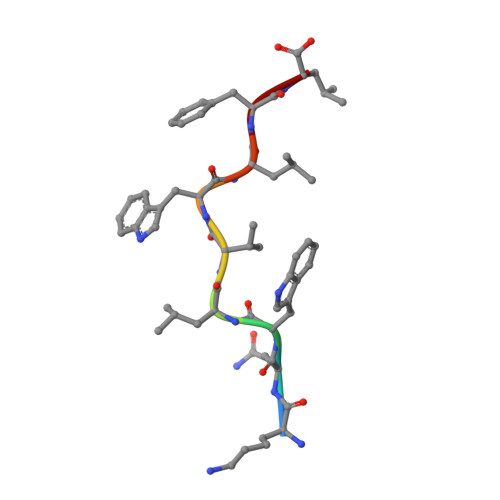
Structural dissimilarity from self drives neoepitope escape from immune tolerance.
Devlin, J.R., Alonso, J.A., Ayres, C.M., Keller, G.L.J., Bobisse, S., Vander Kooi, C.W., Coukos, G., Gfeller, D., Harari, A., Baker, B.M.(2020) Nat Chem Biol 16: 1269-1276
- PubMed: 32807968
- DOI: https://doi.org/10.1038/s41589-020-0610-1
- Primary Citation of Related Structures:
6UJO, 6UJQ, 6UK2, 6UK4 - PubMed Abstract:
T-cell recognition of peptides incorporating nonsynonymous mutations, or neoepitopes, is a cornerstone of tumor immunity and forms the basis of new immunotherapy approaches including personalized cancer vaccines. Yet as they are derived from self-peptides, the means through which immunogenic neoepitopes overcome immune self-tolerance are often unclear. Here we show that a point mutation in a non-major histocompatibility complex anchor position induces structural and dynamic changes in an immunologically active ovarian cancer neoepitope. The changes pre-organize the peptide into a conformation optimal for recognition by a neoepitope-specific T-cell receptor, allowing the receptor to bind the neoepitope with high affinity and deliver potent T-cell signals. Our results emphasize the importance of structural and physical changes relative to self in neoepitope immunogenicity. Considered broadly, these findings can help explain some of the difficulties in identifying immunogenic neoepitopes from sequence alone and provide guidance for developing novel, neoepitope-based personalized therapies.
- Department of Chemistry and Biochemistry, University of Notre Dame, Notre Dame, IN, USA.
Organizational Affiliation: