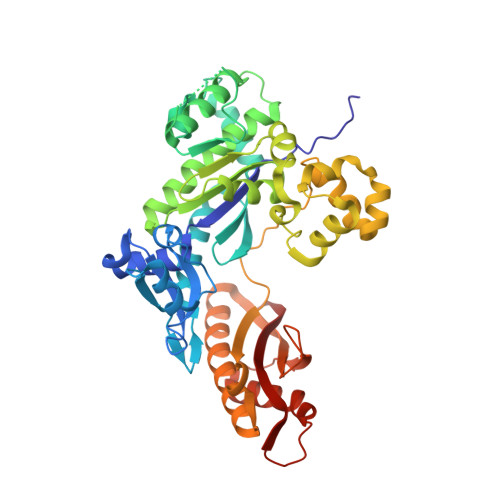
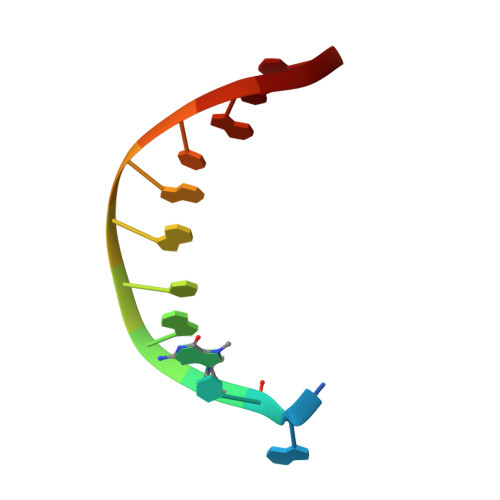
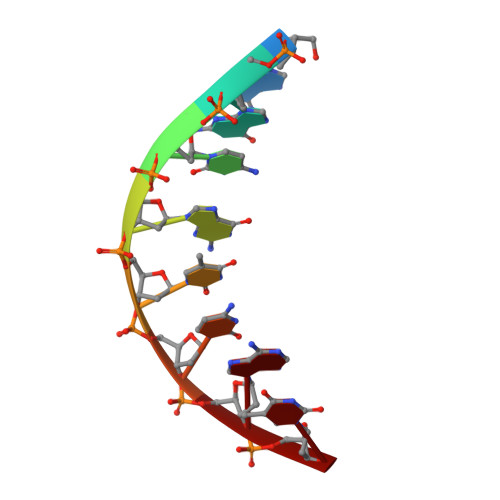
Bypass of the Major Alkylative DNA Lesion by Human DNA Polymerase eta.
Koag, M.C., Jung, H., Kou, Y., Lee, S.(2019) Molecules 24
- PubMed: 31683505
- DOI: https://doi.org/10.3390/molecules24213928
- Primary Citation of Related Structures:
6UI2 - PubMed Abstract:
A wide range of endogenous and exogenous alkylating agents attack DNA to generate various alkylation adducts. N7-methyl-2-deoxyguanosine (Fm7dG) is the most abundant alkylative DNA lesion. If not repaired, Fm7dG can undergo spontaneous depurination, imidazole ring-opening, or bypass by translesion synthesis DNA polymerases. Human DNA polymerase η (polη) efficiently catalyzes across Fm7dG in vitro, but its structural basis is unknown. Herein, we report a crystal structure of polη in complex with templating Fm7dG and an incoming nonhydrolyzable dCTP analog, where a 2'-fluorine-mediated transition destabilization approach was used to prevent the spontaneous depurination of Fm7dG. The structure showed that polη readily accommodated the Fm7dG:dCTP base pair with little conformational change of protein and DNA. In the catalytic site, Fm7dG and dCTP formed three hydrogen bonds with a Watson-Crick geometry, indicating that the major keto tautomer of Fm7dG is involved in base pairing. The polη-Fm7dG:dCTP structure was essentially identical to the corresponding undamaged structure, which explained the efficient bypass of the major methylated lesion. Overall, the first structure of translesion synthesis DNA polymerase bypassing Fm7dG suggests that in the catalytic site of Y-family DNA polymerases, small N7-alkylguanine adducts may be well tolerated and form the canonical Watson-Crick base pair with dCTP through their keto tautomers.
- The Division of Chemical Biology and Medicinal Chemistry, College of Pharmacy, The University of Texas at Austin, 2409 University Avenue, TX 78712, USA. mckoag@gmail.com.
Organizational Affiliation: