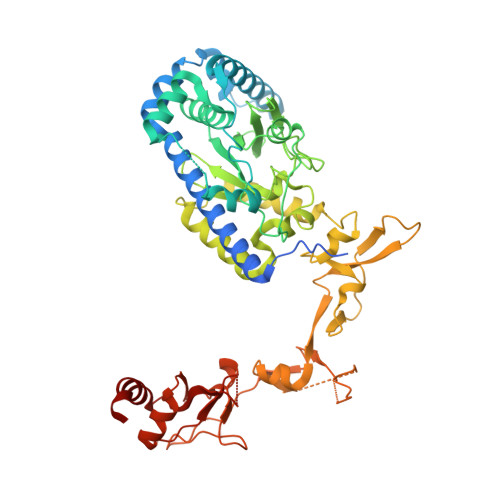
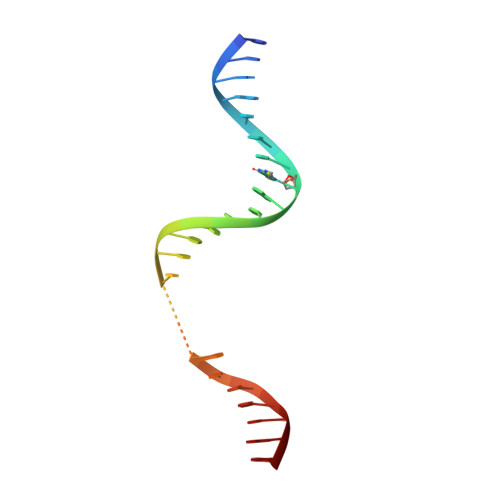
Kinetic gating mechanism of DNA damage recognition by Rad4/XPC.
Chen, X., Velmurugu, Y., Zheng, G., Park, B., Shim, Y., Kim, Y., Liu, L., Van Houten, B., He, C., Ansari, A., Min, J.H.(2015) Nat Commun 6: 5849
- PubMed: 25562780
- DOI: https://doi.org/10.1038/ncomms6849
- Primary Citation of Related Structures:
4YIR, 6UBF - PubMed Abstract:
The xeroderma pigmentosum C (XPC) complex initiates nucleotide excision repair by recognizing DNA lesions before recruiting downstream factors. How XPC detects structurally diverse lesions embedded within normal DNA is unknown. Here we present a crystal structure that captures the yeast XPC orthologue (Rad4) on a single register of undamaged DNA. The structure shows that a disulphide-tethered Rad4 flips out normal nucleotides and adopts a conformation similar to that seen with damaged DNA. Contrary to many DNA repair enzymes that can directly reject non-target sites as structural misfits, our results suggest that Rad4/XPC uses a kinetic gating mechanism whereby lesion selectivity arises from the kinetic competition between DNA opening and the residence time of Rad4/XPC per site. This mechanism is further supported by measurements of Rad4-induced lesion-opening times using temperature-jump perturbation spectroscopy. Kinetic gating may be a general mechanism used by site-specific DNA-binding proteins to minimize time-consuming interrogations of non-target sites.
- Department of Chemistry, University of Illinois at Chicago, 845 W. Taylor Street, Chicago, Illinois 60607, USA.
Organizational Affiliation: