Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation.
Koirala, D., Lewicka, A., Koldobskaya, Y., Huang, H., Piccirilli, J.A.(2020) ACS Chem Biol 15: 205-216
- PubMed: 31765566
- DOI: https://doi.org/10.1021/acschembio.9b00785
- Primary Citation of Related Structures:
6U8D, 6U8K - PubMed Abstract:
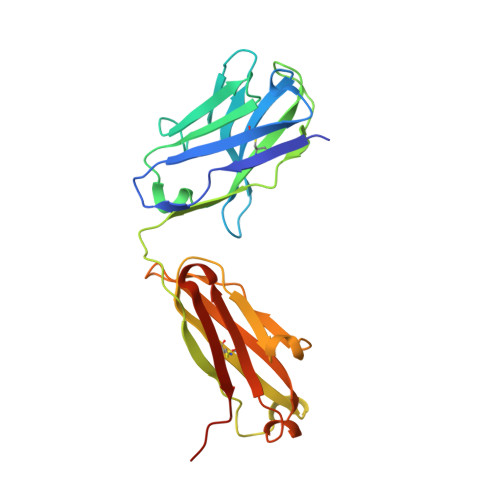
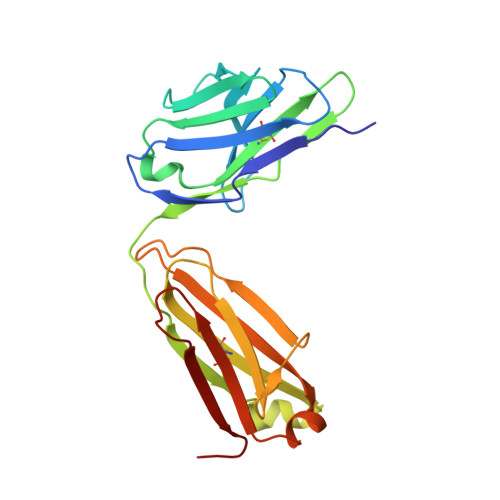
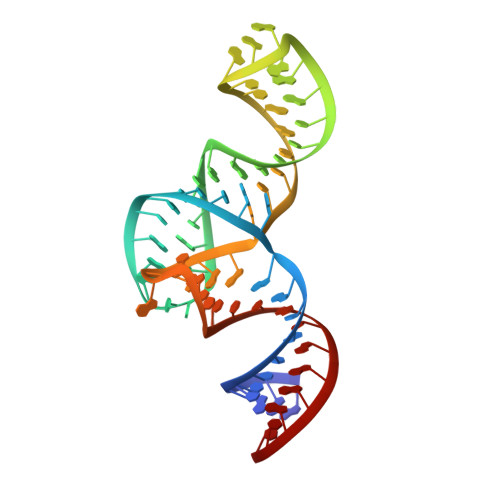
Structured RNA elements within the internal ribosome entry site (IRES) of hepatitis C virus (HCV) genome hijack host cell machinery for translation initiation through a cap-independent mechanism. Here, using a phage display selection, we obtained two antibody fragments (Fabs), HCV2 and HCV3, against HCV IRES that bind the RNA with dissociation constants of 32 ± 7 nM and 37 ± 8 nM respectively, specifically recognizing the so-called junction IIIabc (JIIIabc). We used these Fabs as crystallization chaperones and determined the high-resolution crystal structures of JIIIabc-HCV2 and -HCV3 complexes at 1.81 Å and 2.75 Å resolution respectively, revealing an antiparallel four-way junction with the IIIa and IIIc subdomains brought together through tertiary interactions. The RNA conformation observed in the structures supports the structural model for this region derived from cryo-EM data for the HCV IRES-40S ribosome complex, suggesting that the tertiary fold of the RNA preorganizes the domain for interactions with the 40S ribosome. Strikingly, both Fabs and the ribosomal protein eS27 not only interact with a common subset of nucleotides within the JIIIabc but also use physiochemically similar sets of protein residues to do so, suggesting that the RNA surface is well-suited for interactions with proteins, perhaps analogous to the "hot spot" concept elaborated for protein-protein interactions. Using a rabbit reticulocyte lysate-based translation assay with a bicistronic reporter construct, we further demonstrated that Fabs HCV2 and HCV3 specifically inhibit the HCV IRES-directed translation, implicating disruption of the JIIIabc-ribosome interaction as a potential therapeutic strategy against HCV.
- Department of Biochemistry and Molecular Biology , The University of Chicago , Chicago , Illinois 60637 , United States.
Organizational Affiliation: