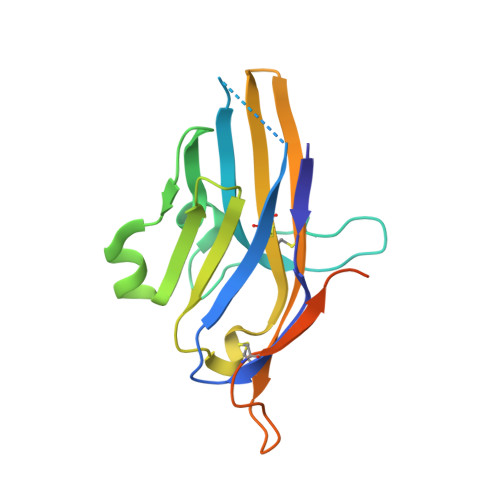
Structural insight into T cell coinhibition by PD-1H (VISTA).
Slater, B.T., Han, X., Chen, L., Xiong, Y.(2020) Proc Natl Acad Sci U S A 117: 1648-1657
- PubMed: 31919279
- DOI: https://doi.org/10.1073/pnas.1908711117
- Primary Citation of Related Structures:
6U6V - PubMed Abstract:
Programmed death-1 homolog (PD-1H), a CD28/B7 family molecule, coinhibits T cell activation and is an attractive immunotherapeutic target for cancer and inflammatory diseases. The molecular basis of its function, however, is unknown. Bioinformatic analyses indicated that PD-1H has a very long Ig variable region (IgV)-like domain and extraordinarily high histidine content, suggesting that unique structural features may contribute to coinhibitory mechanisms. Here we present the 1.9-Å crystal structure of the human PD-1H extracellular domain. It reveals an elongated CC' loop and a striking concentration of histidine residues, located in the complementarity-determining region-like proximal half of the molecule. We show that surface-exposed histidine clusters are essential for robust inhibition of T cell activation. PD-1H exhibits a noncanonical IgV-like topology including an extra "H" β-strand and "clamping" disulfide, absent in known IgV-like structures, that likely restricts its orientation on the cell surface differently from other IgV-like domains. These results provide important insight into a molecular basis of T cell coinhibition by PD-1H.
- Department of Immunobiology, Yale University, New Haven, CT 06511.
Organizational Affiliation: