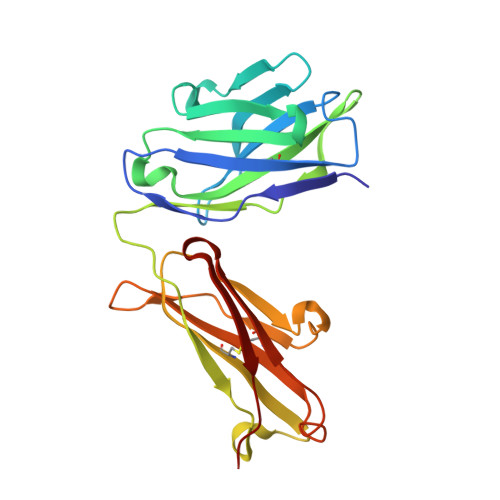
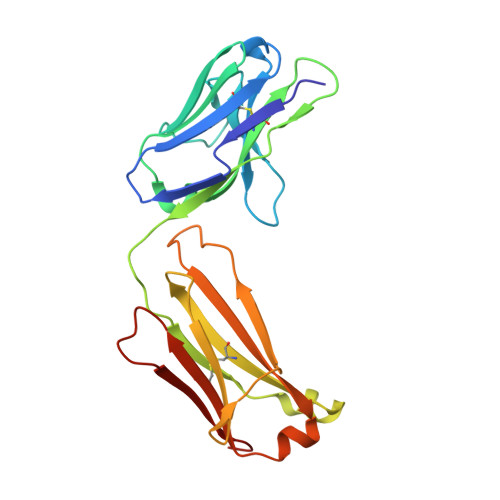
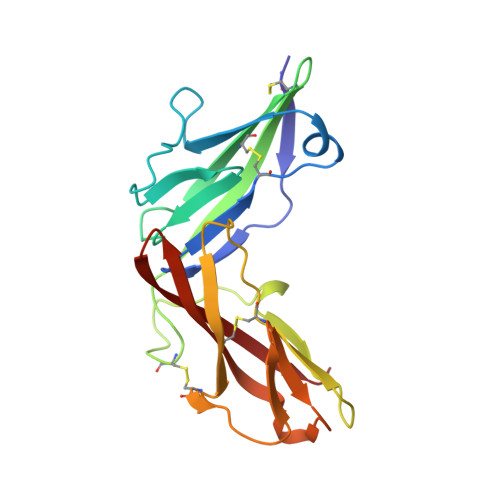
X-ray crystal structure localizes the mechanism of inhibition of an IL-36R antagonist monoclonal antibody to interaction with Ig1 and Ig2 extra cellular domains.
Larson, E.T., Brennan, D.L., Hickey, E.R., Ganesan, R., Kroe-Barrett, R., Farrow, N.A.(2020) Protein Sci 29: 1679-1686
- PubMed: 32239732
- DOI: https://doi.org/10.1002/pro.3862
- Primary Citation of Related Structures:
6U6U - PubMed Abstract:
Cellular signaling via binding of the cytokines IL-36α, β, and γ along with binding of the accessory protein IL-36RAcP, to their cognate receptor IL-36R is believed to play a major role in epithelial and immune cell-mediated inflammation responses. Antagonizing the signaling cascade that results from these binding events via a directed monoclonal antibody provides an opportunity to suppress such immune responses. We report here the molecular structure of a complex between an extracellular portion of human IL-36R and a Fab derived from a high affinity anti-IL-36R neutralizing monoclonal antibody at 2.3 Å resolution. This structure, the first of IL-36R, reveals similarities with other structurally characterized IL-1R family members and elucidates the molecular determinants leading to the high affinity binding of the monoclonal antibody. The structure of the complex reveals that the epitope recognized by the Fab is remote from both the putative ligand and accessory protein binding interfaces on IL-36R, suggesting that the functional activity of the antibody is noncompetitive for these binding events.
- Rheos Medicines, Cambridge, Massachusetts, USA.
Organizational Affiliation: