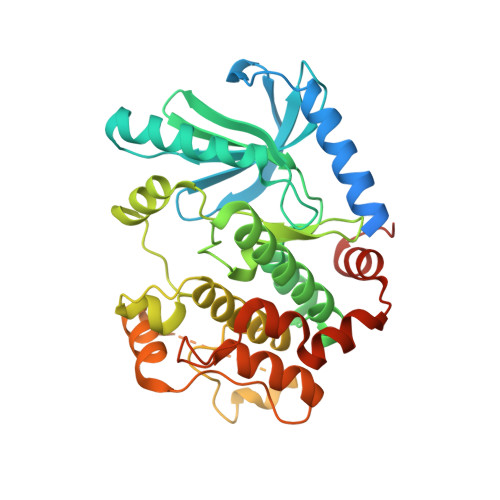
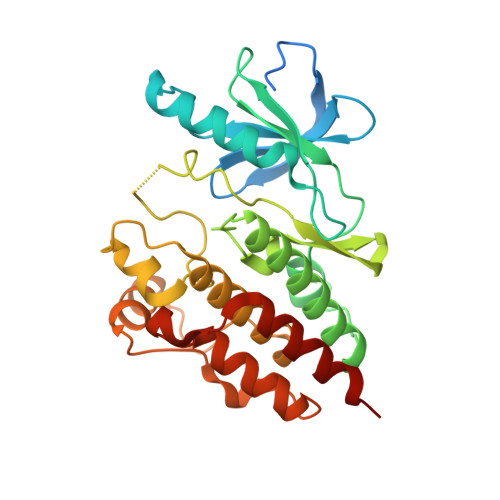
Negative regulation of RAF kinase activity by ATP is overcome by 14-3-3-induced dimerization.
Liau, N.P.D., Wendorff, T.J., Quinn, J.G., Steffek, M., Phung, W., Liu, P., Tang, J., Irudayanathan, F.J., Izadi, S., Shaw, A.S., Malek, S., Hymowitz, S.G., Sudhamsu, J.(2020) Nat Struct Mol Biol 27: 134-141
- PubMed: 31988522
- DOI: https://doi.org/10.1038/s41594-019-0365-0
- Primary Citation of Related Structures:
6U2G, 6U2H - PubMed Abstract:
The RAS-RAF-MEK-ERK signaling axis is frequently activated in human cancers. Physiological concentrations of ATP prevent formation of RAF kinase-domain (RAF KD ) dimers that are critical for activity. Here we present a 2.9-Å-resolution crystal structure of human BRAF KD in complex with MEK and the ATP analog AMP-PCP, revealing interactions between BRAF and ATP that induce an inactive, monomeric conformation of BRAF KD . We also determine how 14-3-3 relieves the negative regulatory effect of ATP through a 2.5-Å-resolution crystal structure of the BRAF KD -14-3-3 complex, in which dimeric 14-3-3 enforces a dimeric BRAF KD assembly to increase BRAF activity. Our data suggest that most oncogenic BRAF mutations alter interactions with ATP and counteract the negative effects of ATP binding by lowering the threshold for RAF dimerization and pathway activation. Our study establishes a framework for rationalizing oncogenic BRAF mutations and provides new avenues for improved RAF-inhibitor discovery.
- Department of Structural Biology, Genentech Inc., South San Francisco, USA.
Organizational Affiliation: