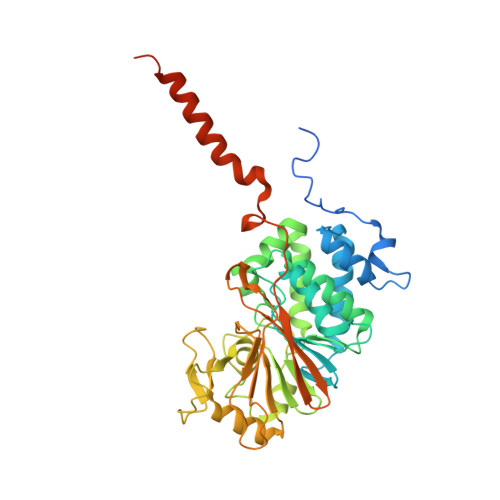
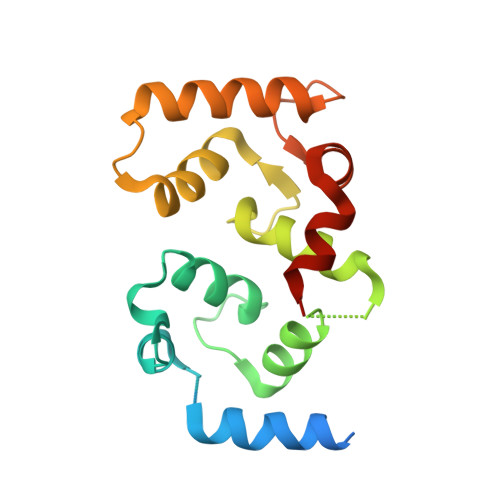
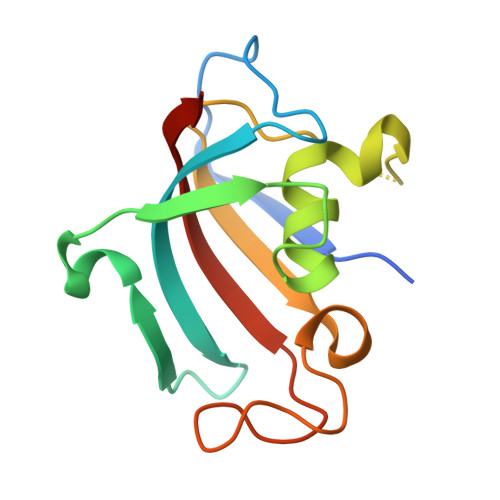
Harnessing calcineurin-FK506-FKBP12 crystal structures from invasive fungal pathogens to develop antifungal agents.
Juvvadi, P.R., Fox 3rd, D., Bobay, B.G., Hoy, M.J., Gobeil, S.M.C., Venters, R.A., Chang, Z., Lin, J.J., Averette, A.F., Cole, D.C., Barrington, B.C., Wheaton, J.D., Ciofani, M., Trzoss, M., Li, X., Lee, S.C., Chen, Y.L., Mutz, M., Spicer, L.D., Schumacher, M.A., Heitman, J., Steinbach, W.J.(2019) Nat Commun 10: 4275-4275
- PubMed: 31537789
- DOI: https://doi.org/10.1038/s41467-019-12199-1
- Primary Citation of Related Structures:
5B8I, 6TZ6, 6TZ7, 6TZ8 - PubMed Abstract:
Calcineurin is important for fungal virulence and a potential antifungal target, but compounds targeting calcineurin, such as FK506, are immunosuppressive. Here we report the crystal structures of calcineurin catalytic (CnA) and regulatory (CnB) subunits complexed with FK506 and the FK506-binding protein (FKBP12) from human fungal pathogens (Aspergillus fumigatus, Candida albicans, Cryptococcus neoformans and Coccidioides immitis). Fungal calcineurin complexes are similar to the mammalian complex, but comparison of fungal and human FKBP12 (hFKBP12) reveals conformational differences in the 40s and 80s loops. NMR analysis, molecular dynamic simulations, and mutations of the A. fumigatus CnA/CnB-FK506-FKBP12-complex identify a Phe88 residue, not conserved in hFKBP12, as critical for binding and inhibition of fungal calcineurin. These differences enable us to develop a less immunosuppressive FK506 analog, APX879, with an acetohydrazine substitution of the C22-carbonyl of FK506. APX879 exhibits reduced immunosuppressive activity and retains broad-spectrum antifungal activity and efficacy in a murine model of invasive fungal infection.
- Division of Pediatric Infectious Diseases, Department of Pediatrics, Duke University Medical Center, Durham, NC, 27710, USA. praveen.juvvadi@duke.edu.
Organizational Affiliation: