Isolation and Structure of an Antibody that Fully Neutralizes Isolate SIVmac239 Reveals Functional Similarity of SIV and HIV Glycan Shields.
Gorman, J., Mason, R.D., Nettey, L., Cavett, N., Chuang, G.Y., Peng, D., Tsybovsky, Y., Verardi, R., Nguyen, R., Ambrozak, D., Biris, K., LaBranche, C.C., Ramesh, A., Schramm, C.A., Zhou, J., Bailer, R.T., Kepler, T.B., Montefiori, D.C., Shapiro, L., Douek, D.C., Mascola, J.R., Roederer, M., Kwong, P.D.(2019) Immunity 51: 724
- PubMed: 31586542
- DOI: https://doi.org/10.1016/j.immuni.2019.09.007
- Primary Citation of Related Structures:
6TYB - PubMed Abstract:
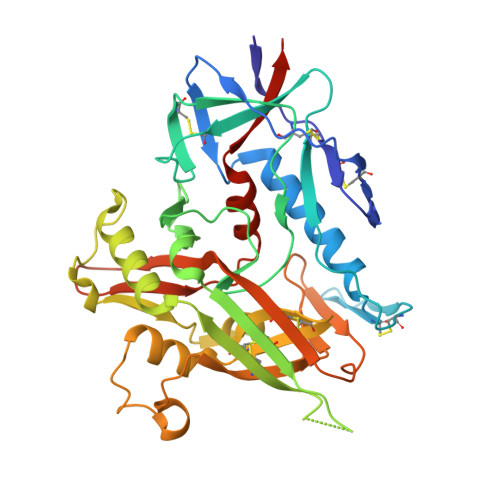
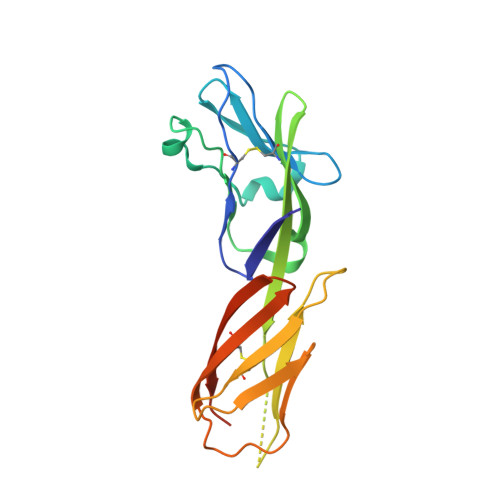
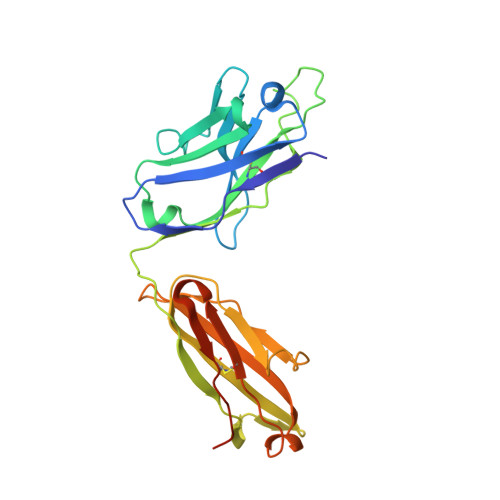
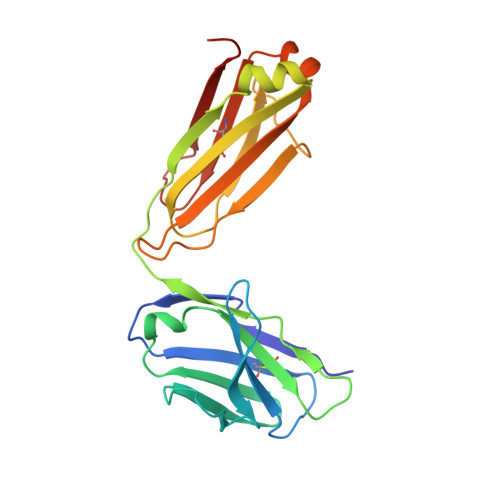
HIV- and SIV-envelope (Env) trimers are both extensively glycosylated, and antibodies identified to date have been unable to fully neutralize SIV mac239 . Here, we report the isolation, structure, and glycan interactions of antibody ITS90.03, a monoclonal antibody that completely neutralized the highly neutralization-resistant isolate, SIV mac239 . The co-crystal structure of a fully glycosylated SIV mac239 -gp120 core in complex with rhesus CD4 and the antigen-binding fragment of ITS90.03 at 2.5-Å resolution revealed that ITS90 recognized an epitope comprised of 45% glycan. SIV-gp120 core, rhesus CD4, and their complex could each be aligned structurally to their human counterparts. The structure revealed that glycans masked most of the SIV Env protein surface, with ITS90 targeting a glycan hole, which is occupied in ∼83% of SIV strains by glycan N238. Overall, the SIV glycan shield appears to functionally resemble its HIV counterpart in coverage of spike, shielding from antibody, and modulation of receptor accessibility.
- Vaccine Research Center, National Institutes of Health, Bethesda, MD 20892, USA.
Organizational Affiliation: