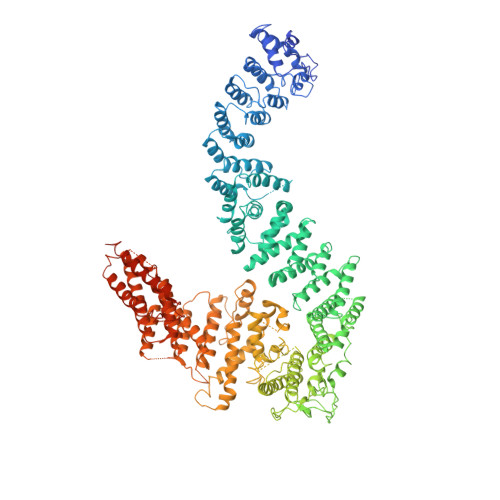
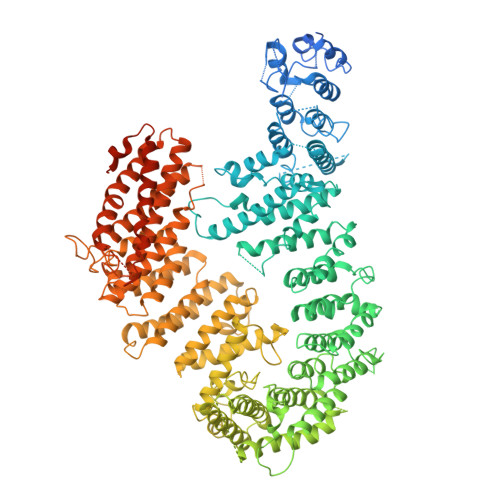
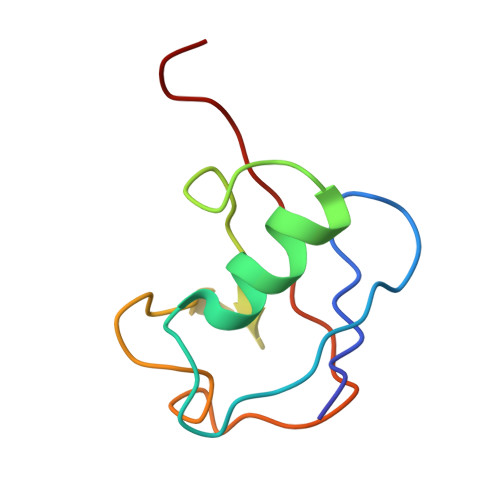
FANCD2-FANCI is a clamp stabilized on DNA by monoubiquitination of FANCD2 during DNA repair.
Alcon, P., Shakeel, S., Chen, Z.A., Rappsilber, J., Patel, K.J., Passmore, L.A.(2020) Nat Struct Mol Biol 27: 240-248
- PubMed: 32066963
- DOI: https://doi.org/10.1038/s41594-020-0380-1
- Primary Citation of Related Structures:
6TNF, 6TNG, 6TNI - PubMed Abstract:
Vertebrate DNA crosslink repair excises toxic replication-blocking DNA crosslinks. Numerous factors involved in crosslink repair have been identified, and mutations in their corresponding genes cause Fanconi anemia (FA). A key step in crosslink repair is monoubiquitination of the FANCD2-FANCI heterodimer, which then recruits nucleases to remove the DNA lesion. Here, we use cryo-EM to determine the structures of recombinant chicken FANCD2 and FANCI complexes. FANCD2-FANCI adopts a closed conformation when the FANCD2 subunit is monoubiquitinated, creating a channel that encloses double-stranded DNA (dsDNA). Ubiquitin is positioned at the interface of FANCD2 and FANCI, where it acts as a covalent molecular pin to trap the complex on DNA. In contrast, isolated FANCD2 is a homodimer that is unable to bind DNA, suggestive of an autoinhibitory mechanism that prevents premature activation. Together, our work suggests that FANCD2-FANCI is a clamp that is locked onto DNA by ubiquitin, with distinct interfaces that may recruit other DNA repair factors.
- MRC Laboratory of Molecular Biology, Cambridge, UK.
Organizational Affiliation: