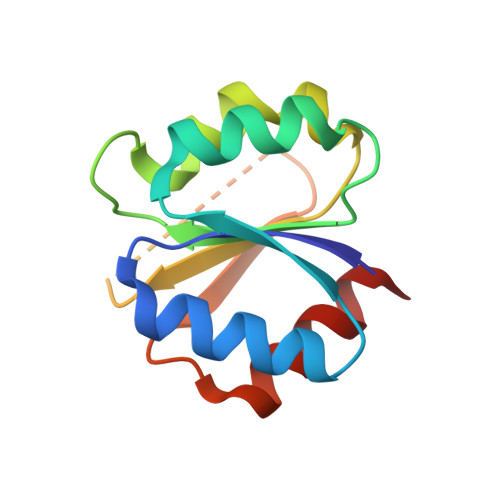
Structural features discriminating hybrid histidine kinase Rec domains from response regulator homologs.
Bruderlin, M., Bohm, R., Fadel, F., Hiller, S., Schirmer, T., Dubey, B.N.(2023) Nat Commun 14: 1002-1002
- PubMed: 36864019
- DOI: https://doi.org/10.1038/s41467-023-36597-8
- Primary Citation of Related Structures:
6TNE - PubMed Abstract:
In two-component systems, the information gathered by histidine kinases (HKs) are relayed to cognate response regulators (RRs). Thereby, the phosphoryl group of the auto-phosphorylated HK is transferred to the receiver (Rec) domain of the RR to allosterically activate its effector domain. In contrast, multi-step phosphorelays comprise at least one additional Rec (Rec inter ) domain that is typically part of the HK and acts as an intermediary for phosphoryl-shuttling. While RR Rec domains have been studied extensively, little is known about discriminating features of Rec inter domains. Here we study the Rec inter domain of the hybrid HK CckA by X-ray crystallography and NMR spectroscopy. Strikingly, all active site residues of the canonical Rec-fold are pre-arranged for phosphoryl-binding and BeF 3 - binding does not alter secondary or quaternary structure, indicating the absence of allosteric changes, the hallmark of RRs. Based on sequence-covariation and modeling, we analyze the intra-molecular DHp/Rec association in hybrid HKs.
- Structural Biology, Biozentrum, University of Basel, Spitalstr. 41, 4056, Basel, Switzerland.
Organizational Affiliation: