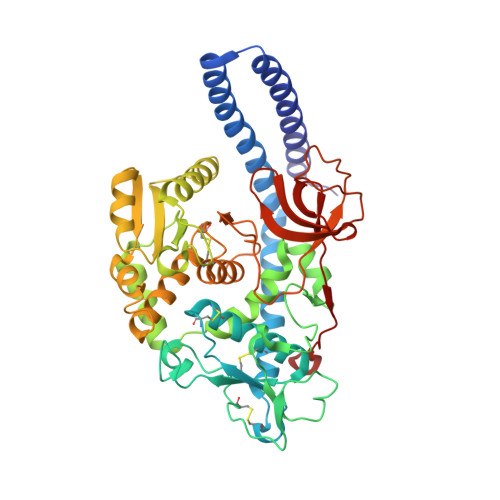
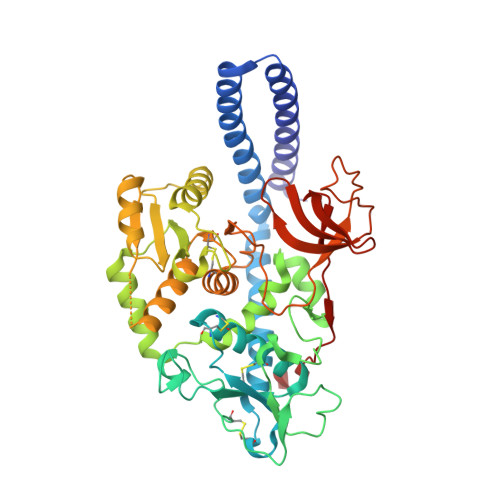
Structural basis for substrate specificity and catalysis of alpha 1,6-fucosyltransferase.
Garcia-Garcia, A., Ceballos-Laita, L., Serna, S., Artschwager, R., Reichardt, N.C., Corzana, F., Hurtado-Guerrero, R.(2020) Nat Commun 11: 973-973
- PubMed: 32080177
- DOI: https://doi.org/10.1038/s41467-020-14794-z
- Primary Citation of Related Structures:
6TKV - PubMed Abstract:
Core-fucosylation is an essential biological modification by which a fucose is transferred from GDP-β-L-fucose to the innermost N-acetylglucosamine residue of N-linked glycans. A single human enzyme α1,6-fucosyltransferase (FUT8) is the only enzyme responsible for this modification via the addition of an α-1,6-linked fucose to N-glycans. To date, the details of substrate recognition and catalysis by FUT8 remain unknown. Here, we report the crystal structure of FUT8 complexed with GDP and a biantennary complex N-glycan (G0), which provides insight into both substrate recognition and catalysis. FUT8 follows an S N 2 mechanism and deploys a series of loops and an α-helix which all contribute in forming the binding site. An exosite, formed by one of these loops and an SH3 domain, is responsible for the recognition of branched sugars, making contacts specifically to the α1,3 arm GlcNAc, a feature required for catalysis. This information serves as a framework for inhibitor design, and helps to assess its potential as a therapeutic target.
- Institute of Biocomputation and Physics of Complex Systems (BIFI), University of Zaragoza, Mariano Esquillor s/n, Campus Rio Ebro, Edificio I+D, Zaragoza, Spain.
Organizational Affiliation: