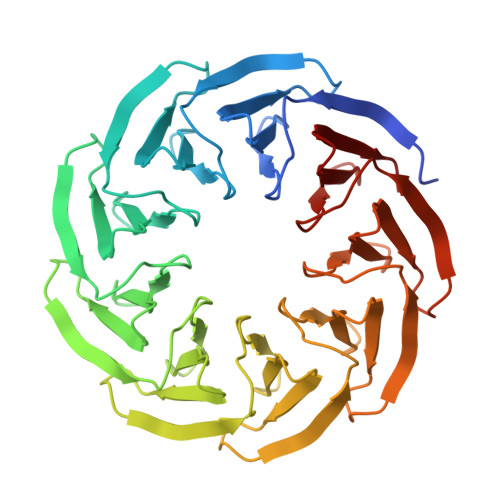
Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Mylemans, B., Laier, I., Kamata, K., Akashi, S., Noguchi, H., Tame, J.R.H., Voet, A.R.D.(2021) FEBS J 288: 530-545
- PubMed: 32343866
- DOI: https://doi.org/10.1111/febs.15347
- Primary Citation of Related Structures:
6TJB, 6TJC, 6TJD, 6TJE, 6TJF, 6TJG, 6TJH, 6TJI - PubMed Abstract:
β-propeller proteins are common in nature, where they are observed to adopt 4- to 10-fold internal rotational pseudo-symmetry. This size diversity can be explained by the evolutionary process of gene duplication and fusion. In this study, we investigated a distorted β-propeller protein, an apparent intermediate between two symmetries. From this template, we created a perfectly symmetric 9-bladed β-propeller named Cake, using computational design and ancestral sequence reconstruction. The designed repeat sequence was found to be capable of generating both 8-fold and 9-fold propellers which are highly stable. Cake variants with 2-10 identical copies of the repeat sequence were characterised by X-ray crystallography and in solution. They were found to be highly stable, and to self-assemble into 8- or 9-fold symmetrical propellers. These findings show that the β-propeller fold allows sufficient structural plasticity to permit a given blade to assemble different forms, a transition from even to odd changes in blade number, and provide a potential explanation for the wide diversity of repeat numbers observed in natural propeller proteins. DATABASE: Structural data are available in Protein Data Bank database under the accession numbers 6TJB, 6TJC, 6TJD, 6TJE, 6TJF, 6TJG, 6TJH and 6TJI.
- Department of Chemistry, KU Leuven, Belgium.
Organizational Affiliation: