Reconstructing the Remote Origins of a Fold Singleton from a Flavodoxin-Like Ancestor.
Toledo-Patino, S., Chaubey, M., Coles, M., Hocker, B.(2019) Biochemistry 58: 4790-4793
- PubMed: 31724394
- DOI: https://doi.org/10.1021/acs.biochem.9b00900
- Primary Citation of Related Structures:
6TH8 - PubMed Abstract:
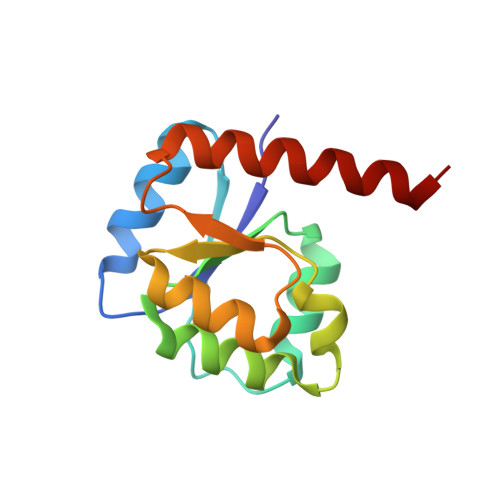
Evolutionary processes that led to the emergence of structured protein domains left footprints in the sequences of modern proteins. We searched for such hints employing state-of-the-art sequence analysis and found evidence that the HemD-like fold emerged from the flavodoxin-like fold through segment swap and gene duplication. To verify this hypothesis, we reverted these evolutionary steps experimentally, constructing a HemD-half that resulted in a protein with the canonical flavodoxin-like architecture. These results of fold reconstruction from the sequence of a different fold strongly support our hypothesis of common ancestry. It further illustrates the plasticity of modern proteins to form new folded proteins.
- Department of Biochemistry , University of Bayreuth , 95447 Bayreuth , Germany.
Organizational Affiliation: