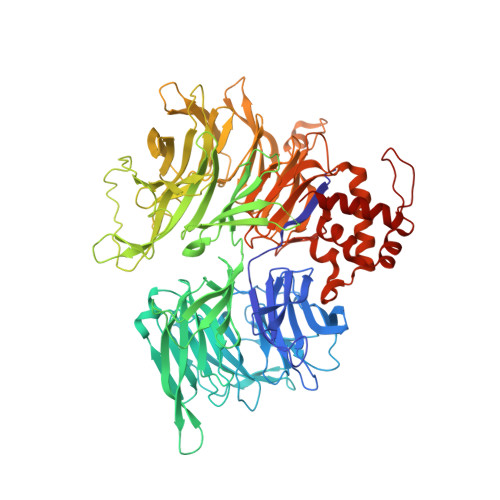
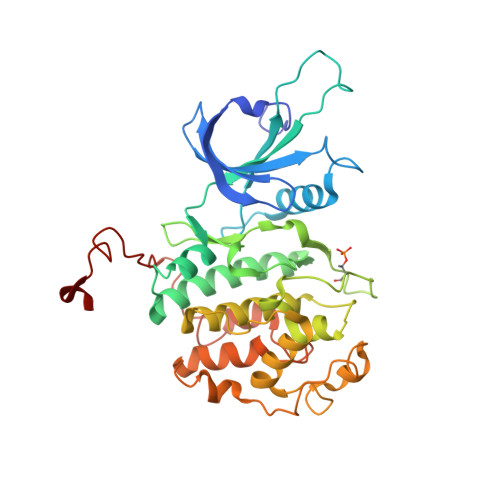
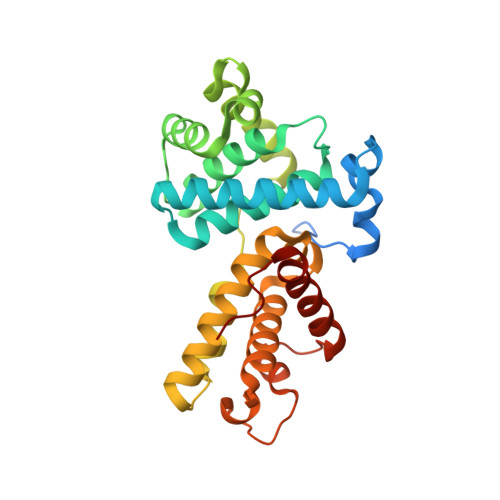
The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K.
Slabicki, M., Kozicka, Z., Petzold, G., Li, Y.D., Manojkumar, M., Bunker, R.D., Donovan, K.A., Sievers, Q.L., Koeppel, J., Suchyta, D., Sperling, A.S., Fink, E.C., Gasser, J.A., Wang, L.R., Corsello, S.M., Sellar, R.S., Jan, M., Gillingham, D., Scholl, C., Frohling, S., Golub, T.R., Fischer, E.S., Thoma, N.H., Ebert, B.L.(2020) Nature 585: 293-297
- PubMed: 32494016
- DOI: https://doi.org/10.1038/s41586-020-2374-x
- Primary Citation of Related Structures:
6TD3 - PubMed Abstract:
Molecular glue compounds induce protein-protein interactions that, in the context of a ubiquitin ligase, lead to protein degradation 1 . Unlike traditional enzyme inhibitors, these molecular glue degraders act substoichiometrically to catalyse the rapid depletion of previously inaccessible targets 2 . They are clinically effective and highly sought-after, but have thus far only been discovered serendipitously. Here, through systematically mining databases for correlations between the cytotoxicity of 4,518 clinical and preclinical small molecules and the expression levels of E3 ligase components across hundreds of human cancer cell lines 3-5 , we identify CR8-a cyclin-dependent kinase (CDK) inhibitor 6 -as a compound that acts as a molecular glue degrader. The CDK-bound form of CR8 has a solvent-exposed pyridyl moiety that induces the formation of a complex between CDK12-cyclin K and the CUL4 adaptor protein DDB1, bypassing the requirement for a substrate receptor and presenting cyclin K for ubiquitination and degradation. Our studies demonstrate that chemical alteration of surface-exposed moieties can confer gain-of-function glue properties to an inhibitor, and we propose this as a broader strategy through which target-binding molecules could be converted into molecular glues.
- Broad Institute of MIT and Harvard, Cambridge, MA, USA.
Organizational Affiliation: