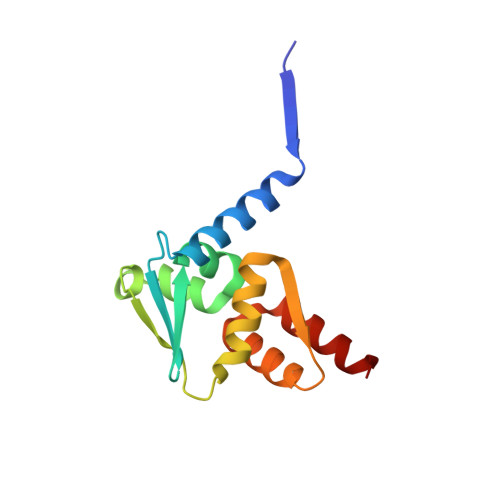
Structural basis of Apt48 inhibition of the BCL6 BTB domain.
Zacharchenko, T., Kalverda, A.P., Wright, S.C.(2021) Structure
- PubMed: 34774129
- DOI: https://doi.org/10.1016/j.str.2021.10.010
- Primary Citation of Related Structures:
6TBT, 6TCJ - PubMed Abstract:
B cell lymphoma 6 (BCL6) is a transcriptional repressor that is deregulated in diffuse large B cell lymphoma, and the peptide aptamer, Apt48, inhibits BCL6 by an unknown mechanism. We report the crystal structure of BCL6 in complex with an Apt48 peptide, and show that Apt48 binds to a therapeutically uncharacterized region at the bottom of the BCL6 BTB domain. We show that the corepressor binding site of the BTB domain may be divided conceptually into two low-affinity, peptide-binding regions. An upper region, the lateral groove, binds peptides in robust three-dimensional conformations, whereas a lower binding site is permissive to less-specific interactions. We show that, even with little sequence specificity, the interactions of the lower region are required for the high-affinity binding of the SMRT corepressor and other peptides to the BTB domain. This has relevance for the design of new BCL6 inhibitors and for understanding the evolution of corepressor interactions with the BTB domain.
- School of Biology and the Astbury Centre for Structural Molecular Biology, University of Leeds, Leeds LS2 9JT, UK.
Organizational Affiliation: