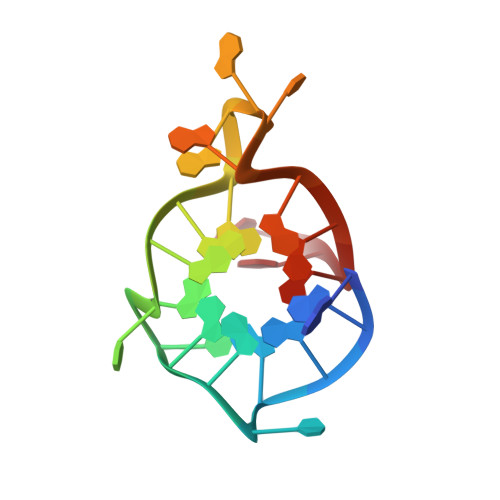
1H,13C, and15N chemical shift assignments of a G-quadruplex forming sequence within the KRAS proto-oncogene promoter region.
Marquevielle, J., Kumar, M.V.V., Mergny, J.L., Salgado, G.F.(2018) Biomol NMR Assign 12: 123-127
- PubMed: 29189986
- DOI: https://doi.org/10.1007/s12104-017-9793-0
- Primary Citation of Related Structures:
6T51 - PubMed Abstract:
Single stranded guanine rich DNA (or RNA) sequences adopt noncanonical secondary structures called G-quadruplexes (G4). Functionally, quadruplexes control gene transcription and regulate activities such as replication, gene recombination or alternative splicing. Hence they are potential targets for cancer, neuronal, and viral related diseases. KRAS is one of the most mutated oncogenes in the genome of cancer cells and contains a nuclease hypersensitive element (NHE) sequence capable of forming G-quadruplexes via its six runs of guanines. In our work, we are interested in the NMR structure of the major G4 scaffold formed in the KRAS NHE region with a mutated sequence of 22 residues. Here, we report 1 H, 13 C and 15 N chemical shift assignments the G4 formed within KRAS22RT sequence.
- ARNA Laboratory, European Institute of Chemistry and Biology (IECB), Université de Bordeaux, Inserm U1212 - CNRS UMR 5320, 2, rue Robert Escarpit, 33607, Pessac, France.
Organizational Affiliation: