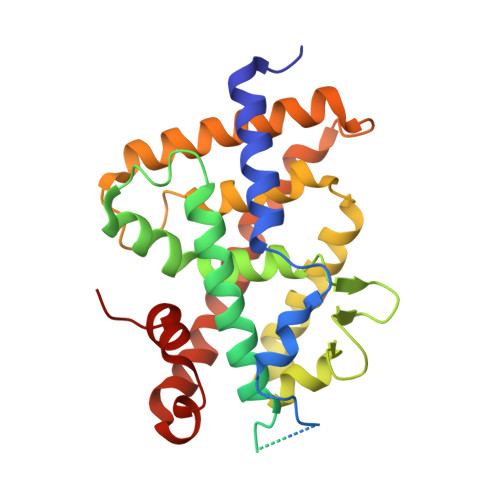
Structural Analysis of VDR Complex with ZK168281 Antagonist.
Belorusova, A.Y., Chalhoub, S., Rovito, D., Rochel, N.(2020) J Med Chem 63: 9457-9463
- PubMed: 32787090
- DOI: https://doi.org/10.1021/acs.jmedchem.0c00656
- Primary Citation of Related Structures:
6T2M - PubMed Abstract:
Vitamin D receptor (VDR) antagonists prevent the VDR activation function helix 12 from folding into its active conformation, thus affecting coactivator recruitment and antagonizing the transcriptional regulation induced by 1α,25-dihydroxyvitamin D3. Here, we report the crystal structure of the zebrafish VDR ligand-binding domain in complex with the ZK168281 antagonist, revealing that the ligand prevents optimal folding of the C-terminal region of VDR. This interference was confirmed by hydrogen-deuterium exchange mass spectrometry (HDX-MS) in solution.
- Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), 67400 Illkirch, France.
Organizational Affiliation: