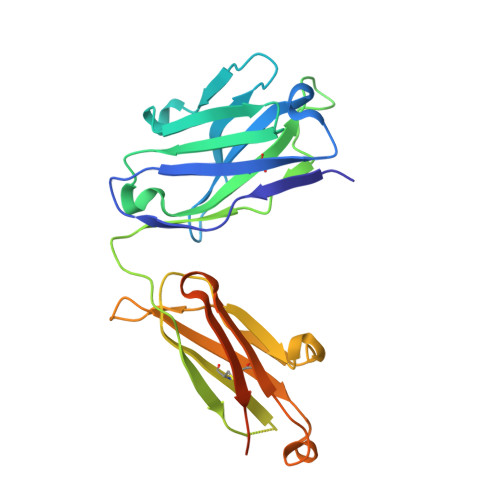
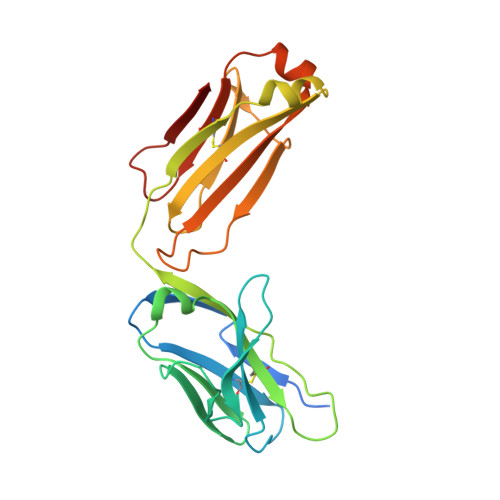
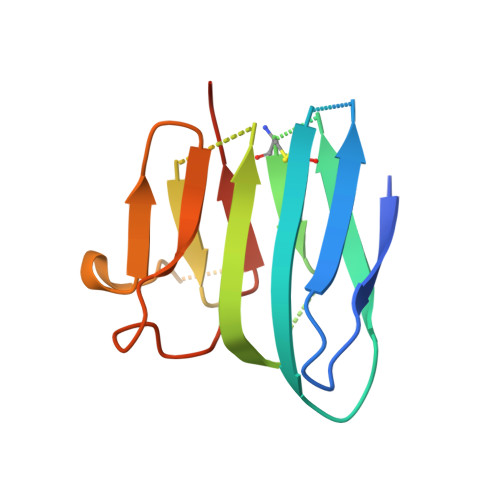
Crystal structure and receptor-interacting residues of MYDGF - a protein mediating ischemic tissue repair.
Ebenhoch, R., Akhdar, A., Reboll, M.R., Korf-Klingebiel, M., Gupta, P., Armstrong, J., Huang, Y., Frego, L., Rybina, I., Miglietta, J., Pekcec, A., Wollert, K.C., Nar, H.(2019) Nat Commun 10: 5379-5379
- PubMed: 31772377
- DOI: https://doi.org/10.1038/s41467-019-13343-7
- Primary Citation of Related Structures:
6SVK, 6SVL - PubMed Abstract:
Myeloid-derived growth factor (MYDGF) is a paracrine-acting protein that is produced by bone marrow-derived monocytes and macrophages to protect and repair the heart after myocardial infarction (MI). This effect can be used for the development of protein-based therapies for ischemic tissue repair, also beyond the sole application in heart tissue. Here, we report the X-ray structure of MYDGF and identify its functionally relevant receptor binding epitope. MYDGF consists of a 10-stranded β-sandwich with a folding topology showing no similarities to other cytokines or growth factors. By characterizing the epitope of a neutralizing antibody and utilizing functional assays to study the activity of surface patch-mutations, we were able to localize the receptor interaction interface to a region around two surface tyrosine residues 71 and 73 and an adjacent prominent loop structure of residues 97-101. These findings enable structure-guided protein engineering to develop modified MYDGF variants with potentially improved properties for clinical use.
- Boehringer Ingelheim Pharma GmbH & Co. KG, Birkendorfer Str. 65, 88397, Biberach an der Riss, Germany.
Organizational Affiliation: