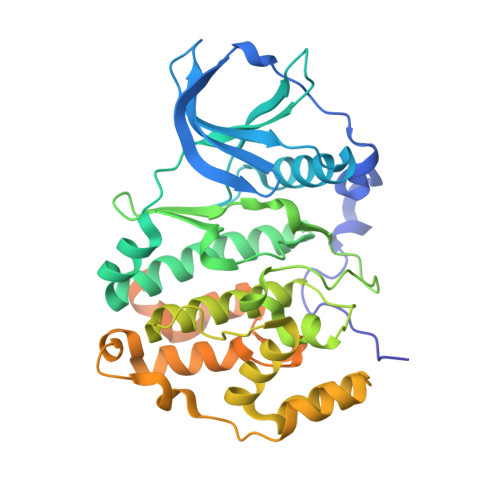
Unexpected CK2 beta-antagonistic functionality of bisubstrate inhibitors targeting protein kinase CK2.
Pietsch, M., Viht, K., Schnitzler, A., Ekambaram, R., Steinkruger, M., Enkvist, E., Nienberg, C., Nickelsen, A., Lauwers, M., Jose, J., Uri, A., Niefind, K.(2020) Bioorg Chem 96: 103608-103608
- PubMed: 32058103
- DOI: https://doi.org/10.1016/j.bioorg.2020.103608
- Primary Citation of Related Structures:
6SPW, 6SPX - PubMed Abstract:
Protein kinase CK2, a heterotetrameric holoenzyme composed of two catalytic chains (CK2α) attached to a homodimer of regulatory subunits (CK2β), is a target for drug development for cancer therapy. Here, we describe the tetraiodobenzimidazole derivative ARC-3140, a bisubstrate inhibitor addressing the ATP site and the substrate-binding site of CK2 with extraordinary affinity (K i = 84 pM). In a crystal structure of ARC-3140 in complex with CK2α, three copies of the inhibitor are visible, one of them at the CK2β interface of CK2α. Subsequent interaction studies based on microscale thermophoresis and fluorescence anisotropy changes revealed a significant impact of ARC-3140 and of its tetrabromo equivalent ARC-1502 on the CK2α/CK2β interaction. A structural inspection revealed that ARC-3140, unlike CK2β antagonists described so far, interferes with both sub-interfaces of the bipartite CK2α/CK2β interaction. Thus, ARC-3140 is a lead for the further development of highly effective compounds perturbating the quaternary structure of the CK2α 2 β 2 holoenzyme.
- Institut II für Pharmakologie, Zentrum für Pharmakologie, Medizinische Fakultät, Universität zu Köln, Gleueler Str. 24, D-50931 Köln, Germany.
Organizational Affiliation: