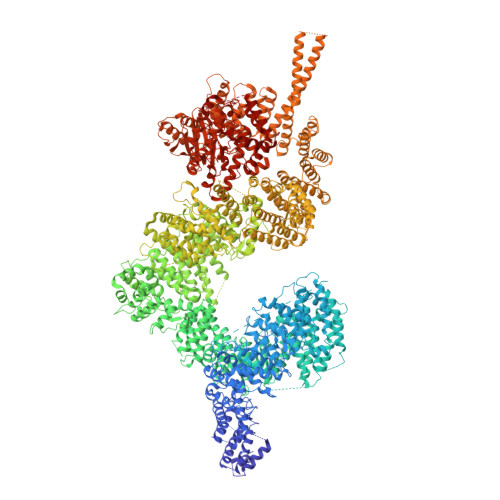
Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Jansma, M., Linke-Winnebeck, C., Eustermann, S., Lammens, K., Kostrewa, D., Stakyte, K., Litz, C., Kessler, B., Hopfner, K.P.(2020) Structure 28: 83-95.e5
- PubMed: 31740028
- DOI: https://doi.org/10.1016/j.str.2019.10.013
- Primary Citation of Related Structures:
6SKY, 6SKZ, 6SL0, 6SL1 - PubMed Abstract:
Tel1 (ATM in humans) is a large kinase that resides in the cell in an autoinhibited dimeric state and upon activation orchestrates the cellular response to DNA damage. We report the structure of an endogenous Tel1 dimer from Chaetomium thermophilum. Major parts are at 2.8 Å resolution, including the kinase active site with ATPγS bound, and two different N-terminal solenoid conformations are at 3.4 Å and 3.6 Å, providing a side-chain model for 90% of the Tel1 polypeptide. We show that the N-terminal solenoid has DNA binding activity, but that its movements are not coupled to kinase activation. Although ATPγS and catalytic residues are poised for catalysis, the kinase resides in an autoinhibited state. The PIKK regulatory domain acts as a pseudo-substrate, blocking direct access to the site of catalysis. The structure allows mapping of human cancer mutations and defines mechanisms of autoinhibition at near-atomic resolution.
- Department of Biochemistry, Ludwig-Maximilians-Universität, Feodor-Lynen-Strasse 25, 81377 Munich, Germany; Gene Center, Ludwig-Maximilians-Universität, Feodor-Lynen-Strasse 25, 81377 Munich, Germany.
Organizational Affiliation: