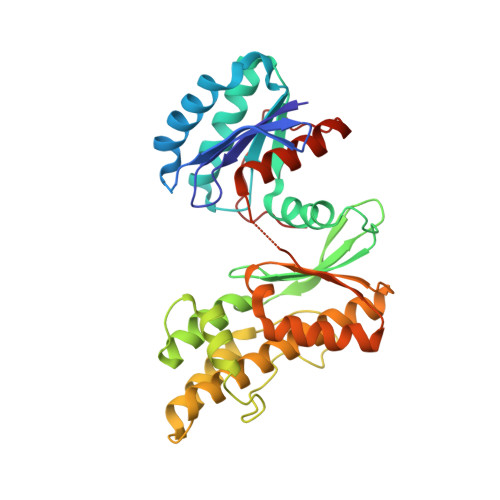
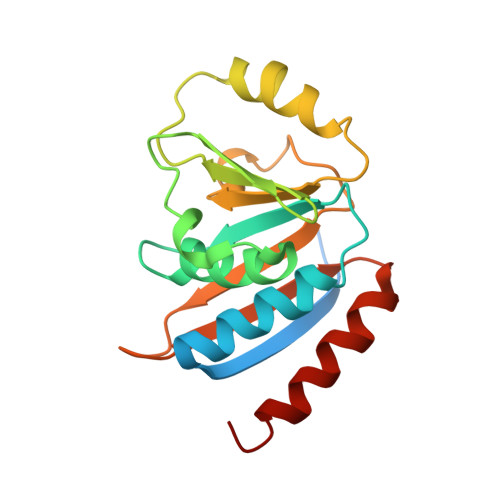
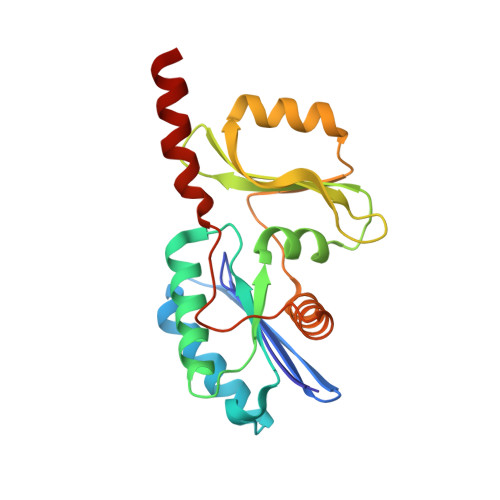
The structure of the TsaB/TsaD/TsaE complex reveals an unexpected mechanism for the bacterial t6A tRNA-modification.
Missoury, S., Plancqueel, S., Li de la Sierra-Gallay, I., Zhang, W., Liger, D., Durand, D., Dammak, R., Collinet, B., van Tilbeurgh, H.(2018) Nucleic Acids Res 46: 5850-5860
- PubMed: 29741707
- DOI: https://doi.org/10.1093/nar/gky323
- Primary Citation of Related Structures:
6S84 - PubMed Abstract:
The universal N6-threonylcarbamoyladenosine (t6A) modification at position A37 of ANN-decoding tRNAs is essential for translational fidelity. In bacteria the TsaC enzyme first synthesizes an l-threonylcarbamoyladenylate (TC-AMP) intermediate. In cooperation with TsaB and TsaE, TsaD then transfers the l-threonylcarbamoyl-moiety from TC-AMP onto tRNA. We determined the crystal structure of the TsaB-TsaE-TsaD (TsaBDE) complex of Thermotoga maritima in presence of a non-hydrolysable AMPCPP. TsaE is positioned at the entrance of the active site pocket of TsaD, contacting both the TsaB and TsaD subunits and prohibiting simultaneous tRNA binding. AMPCPP occupies the ATP binding site of TsaE and is sandwiched between TsaE and TsaD. Unexpectedly, the binding of TsaE partially denatures the active site of TsaD causing loss of its essential metal binding sites. TsaE interferes in a pre- or post-catalytic step and its binding to TsaBD is regulated by ATP hydrolysis. This novel binding mode and activation mechanism of TsaE offers good opportunities for antimicrobial drug development.
- Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS UMR 9198, Univ. Paris-Sud, Université Paris-Saclay, 91198 Gif sur Yvette Cedex, France.
Organizational Affiliation: