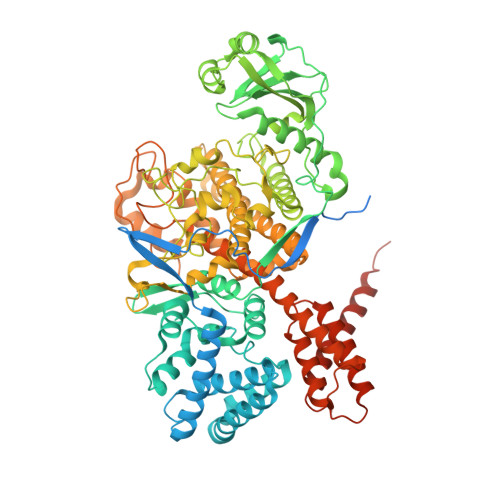
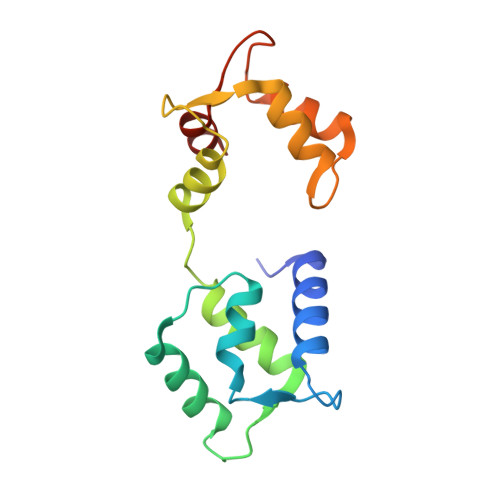
Inhibition of bacterial ubiquitin ligases by SidJ-calmodulin catalysed glutamylation.
Bhogaraju, S., Bonn, F., Mukherjee, R., Adams, M., Pfleiderer, M.M., Galej, W.P., Matkovic, V., Lopez-Mosqueda, J., Kalayil, S., Shin, D., Dikic, I.(2019) Nature 572: 382-386
- PubMed: 31330532
- DOI: https://doi.org/10.1038/s41586-019-1440-8
- Primary Citation of Related Structures:
6S5T - PubMed Abstract:
The family of bacterial SidE enzymes catalyses phosphoribosyl-linked serine ubiquitination and promotes infectivity of Legionella pneumophila, a pathogenic bacteria that causes Legionnaires' disease 1-3 . SidE enzymes share the genetic locus with the Legionella effector SidJ that spatiotemporally opposes the toxicity of these enzymes in yeast and mammalian cells, through a mechanism that is currently unknown 4-6 . Deletion of SidJ leads to a substantial defect in the growth of Legionella in both its natural hosts (amoebae) and in mouse macrophages 4,5 . Here we demonstrate that SidJ is a glutamylase that modifies the catalytic glutamate in the mono-ADP ribosyl transferase domain of the SdeA, thus blocking the ubiquitin ligase activity of SdeA. The glutamylation activity of SidJ requires interaction with the eukaryotic-specific co-factor calmodulin, and can be regulated by intracellular changes in Ca 2+ concentrations. The cryo-electron microscopy structure of SidJ in complex with human apo-calmodulin revealed the architecture of this heterodimeric glutamylase. We show that, in cells infected with L. pneumophila, SidJ mediates the glutamylation of SidE enzymes on the surface of vacuoles that contain Legionella. We used quantitative proteomics to uncover multiple host proteins as putative targets of SidJ-mediated glutamylation. Our study reveals the mechanism by which SidE ligases are inhibited by a SidJ-calmodulin glutamylase, and opens avenues for exploring an understudied protein modification (glutamylation) in eukaryotes.
- Institute of Biochemistry II, Faculty of Medicine, Goethe University, Frankfurt am Main, Germany. bhogaraju@embl.fr.
Organizational Affiliation: