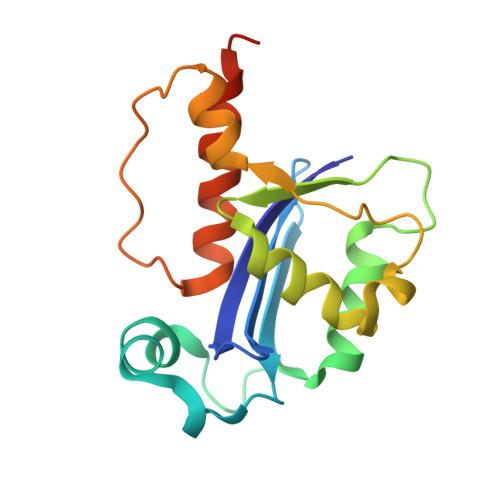
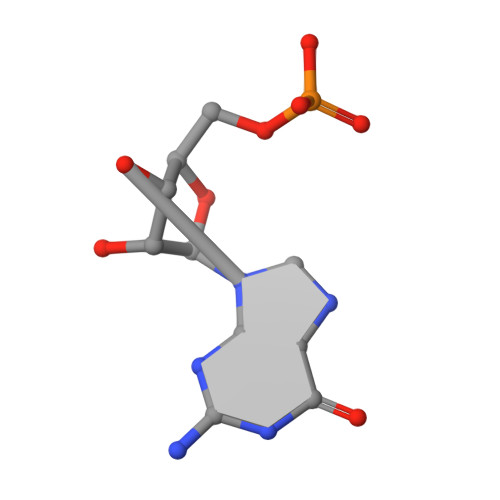
Structural and dynamic studies provide insights into specificity and allosteric regulation of ribonuclease as, a key enzyme in mycobacterial virulence.
Calvanese, L., Squeglia, F., Romano, M., D'Auria, G., Falcigno, L., Berisio, R.(2020) J Biomol Struct Dyn 38: 2455-2467
- PubMed: 31299874
- DOI: https://doi.org/10.1080/07391102.2019.1643786
- Primary Citation of Related Structures:
6S0M - PubMed Abstract:
Ribonuclease AS (RNase AS) is a crucial enzyme for virulence of Mycobacterium tuberculosis . We previously observed that RNase AS structurally resembles RNase T from Escherichia coli , an important enzyme for tRNA maturation and turnover. Here, we combine X-ray crystallography and molecular dynamics (MD) to investigate the specificity and dynamic properties of substrate binding. Both X-ray and MD data provide structural determinants that corroborate the strict substrate specificity of RNase AS to cleave only adenosine residues, due to the structural features of adenine base. Beside suggesting tRNA as most likely substrate of RNase AS, MD and modeling studies identify key enzyme-ligand interactions, both involving the catalytic site and the double helix region of tRNA, which is locked by interactions with a set of arginine residues. The MD data also evidence a ligand-induced conformational change of the enzyme which is transferred from one chain to the adjacent one. These data will explain the dimeric nature of both RNase AS and RNase T, with two catalytic grooves composed of both chains. Also, they account for the dichotomy of tRNA, which contains both the substrate poly(A) chain and an inhibiting double strand RNA. Indeed, they provide a possible mechanism of allosteric regulation, which unlocks one catalytic groove when the second groove is inhibited by the double strand region of tRNA. Finally, a full comprehension of the molecular details of tRNA maturation processes is essential to develop novel strategies to modulate RNA processing, for therapeutic purposes. AbbreviationsMDmolecular dynamicsPDBProtein Data BankRMSDroot mean square deviationRMSFroot mean square fluctuationRNAribonucleotidic acidRNase ASRibonuclease ASCommunicated by Ramasamy H. Sarma.
- CIRPeB, University of Naples Federico II, Naples, Italy.
Organizational Affiliation: