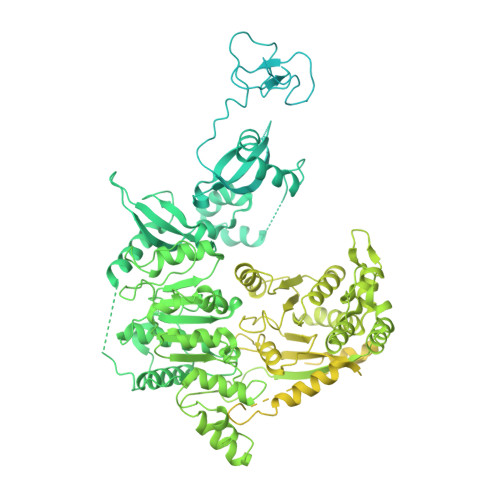
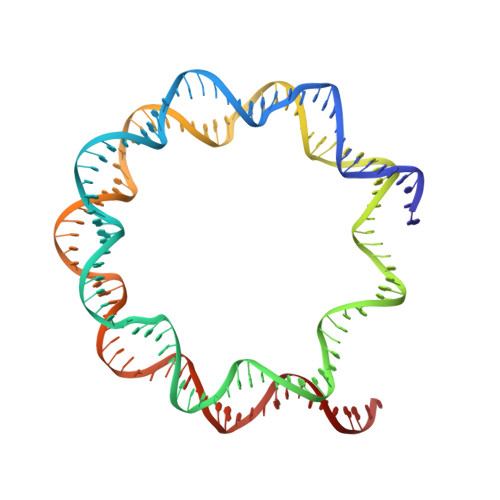
Nucleosome-CHD4 chromatin remodeller structure maps human disease mutations.
Farnung, L., Ochmann, M., Cramer, P.(2020) Elife 9
- PubMed: 32543371
- DOI: https://doi.org/10.7554/eLife.56178
- Primary Citation of Related Structures:
6RYR, 6RYU - PubMed Abstract:
Chromatin remodeling plays important roles in gene regulation during development, differentiation and in disease. The chromatin remodeling enzyme CHD4 is a component of the NuRD and ChAHP complexes that are involved in gene repression. Here, we report the cryo-electron microscopy (cryo-EM) structure of Homo sapiens CHD4 engaged with a nucleosome core particle in the presence of the non-hydrolysable ATP analogue AMP-PNP at an overall resolution of 3.1 Å. The ATPase motor of CHD4 binds and distorts nucleosomal DNA at superhelical location (SHL) +2, supporting the 'twist defect' model of chromatin remodeling. CHD4 does not induce unwrapping of terminal DNA, in contrast to its homologue Chd1, which functions in gene activation. Our structure also maps CHD4 mutations that are associated with human cancer or the intellectual disability disorder Sifrim-Hitz-Weiss syndrome.
- Max Planck Institute for Biophysical Chemistry, Department of Molecular Biology, Göttingen, Germany.
Organizational Affiliation: