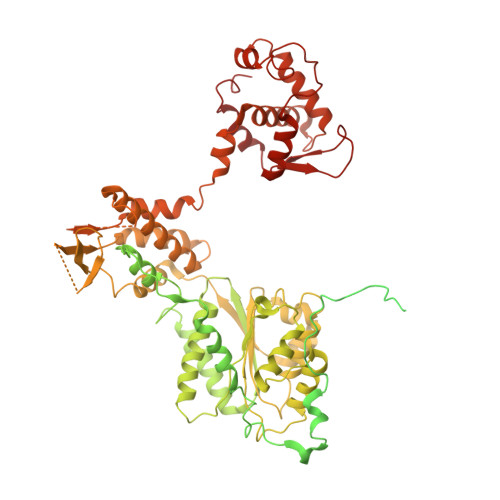
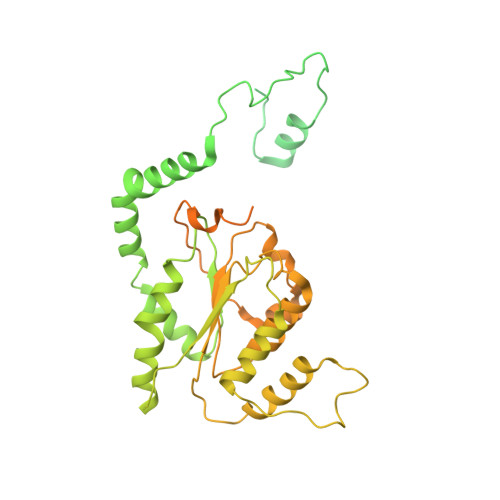
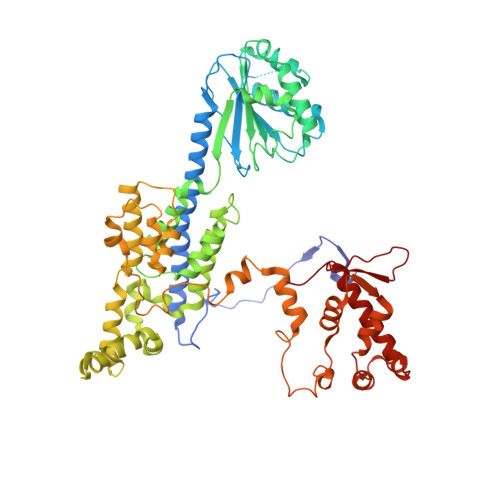
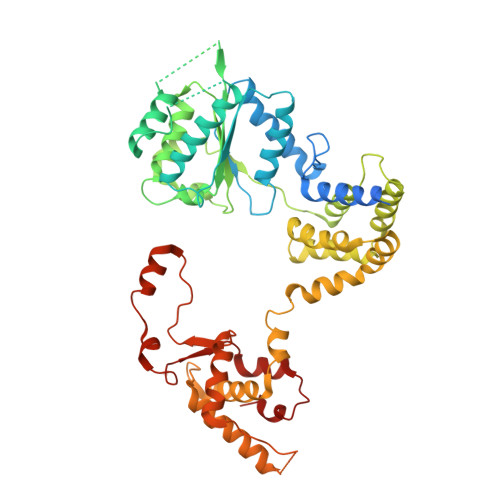
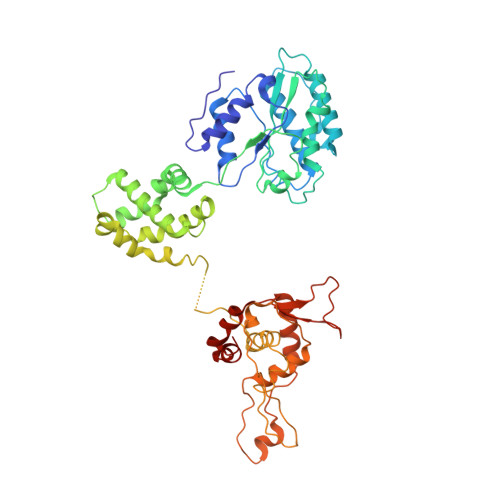
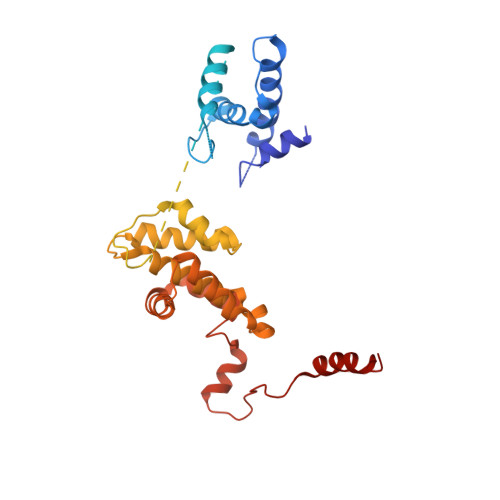
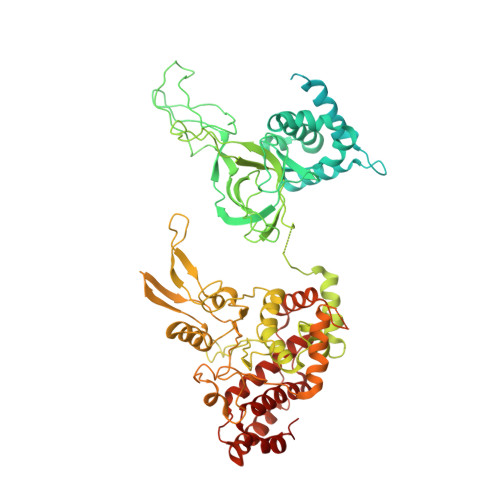
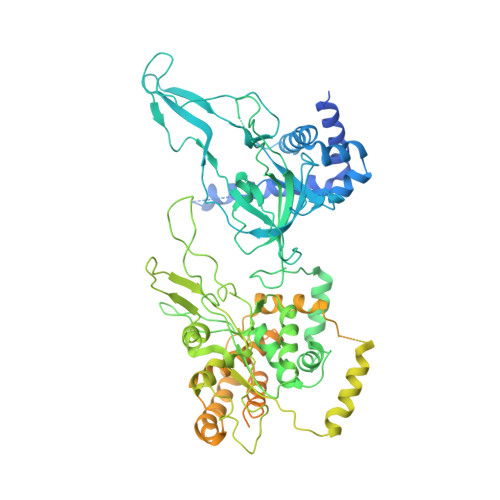
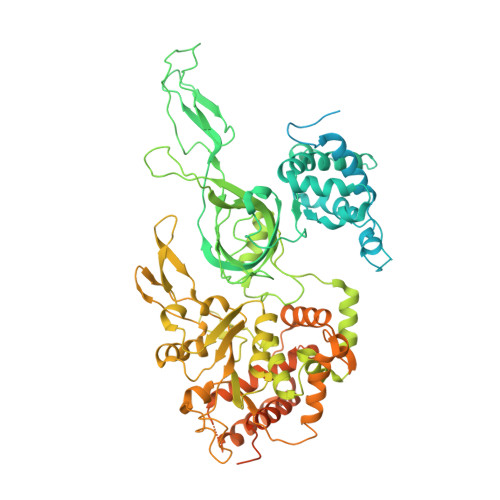
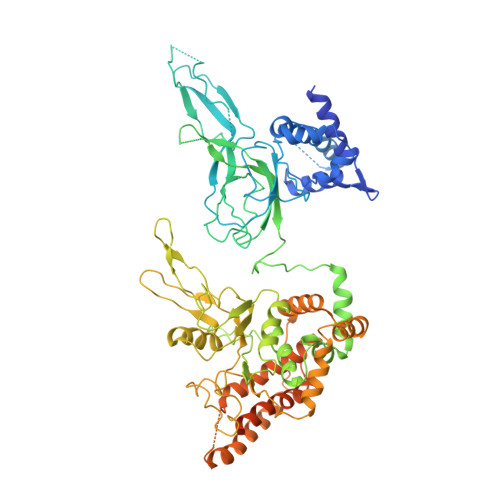
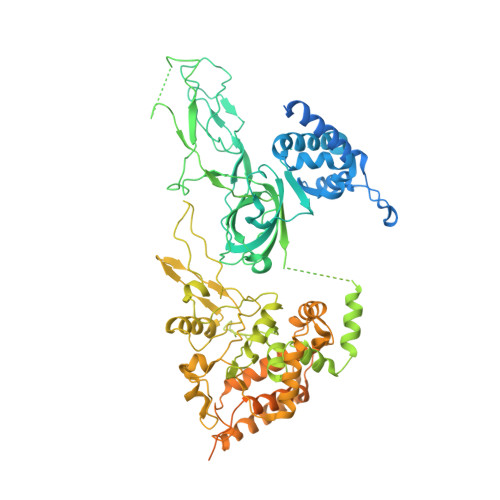
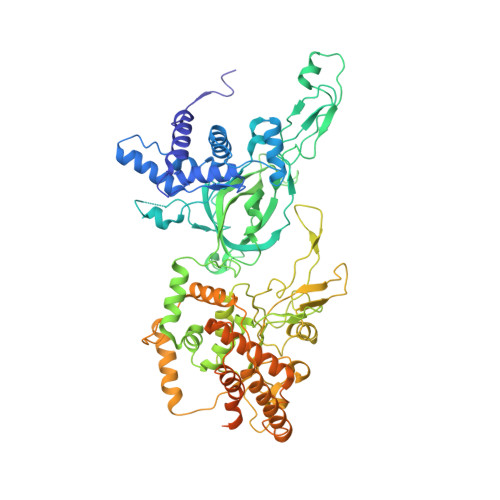
Mechanism of head-to-head MCM double-hexamer formation revealed by cryo-EM.
Miller, T.C.R., Locke, J., Greiwe, J.F., Diffley, J.F.X., Costa, A.(2019) Nature 575: 704-710
- PubMed: 31748745
- DOI: https://doi.org/10.1038/s41586-019-1768-0
- Primary Citation of Related Structures:
6RQC - PubMed Abstract:
In preparation for bidirectional DNA replication, the origin recognition complex (ORC) loads two hexameric MCM helicases to form a head-to-head double hexamer around DNA 1,2 . The mechanism of MCM double-hexamer formation is debated. Single-molecule experiments have suggested a sequential mechanism, in which the ORC-dependent loading of the first hexamer drives the recruitment of the second hexamer 3 . By contrast, biochemical data have shown that two rings are loaded independently via the same ORC-mediated mechanism, at two inverted DNA sites 4,5 . Here we visualize MCM loading using time-resolved electron microscopy, and identify intermediates in the formation of the double hexamer. We confirm that both hexamers are recruited via the same interaction that occurs between ORC and the C-terminal domains of the MCM helicases. Moreover, we identify the mechanism of coupled MCM loading. The loading of the first MCM hexamer around DNA creates a distinct interaction site, which promotes the engagement of ORC at the N-terminal homodimerization interface of MCM. In this configuration, ORC is poised to direct the recruitment of the second hexamer in an inverted orientation, which is suitable for the formation of the double hexamer. Our results therefore reconcile the two apparently contrasting models derived from single-molecule experiments and biochemical data.
- Macromolecular Machines Laboratory, Francis Crick Institute, London, UK.
Organizational Affiliation: