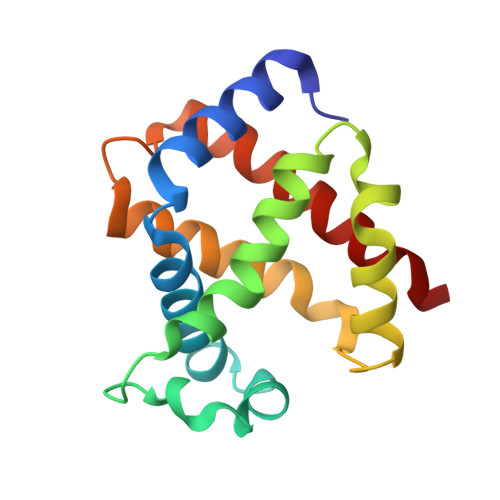
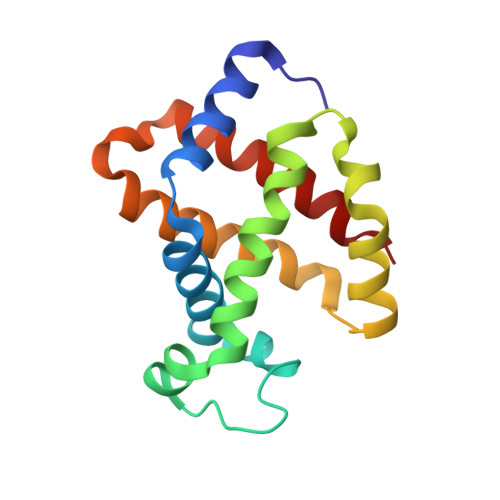
The unique structural features of carbonmonoxy hemoglobin from the sub-Antarctic fish Eleginops maclovinus.
Balasco, N., Vitagliano, L., Merlino, A., Verde, C., Mazzarella, L., Vergara, A.(2019) Sci Rep 9: 18987-18987
- PubMed: 31831781
- DOI: https://doi.org/10.1038/s41598-019-55331-3
- Primary Citation of Related Structures:
6RP5 - PubMed Abstract:
Tetrameric hemoglobins (Hbs) are prototypical systems for the investigations of fundamental properties of proteins. Although the structure of these proteins has been known for nearly sixty years, there are many aspects related to their function/structure that are still obscure. Here, we report the crystal structure of a carbonmonoxy form of the Hb isolated from the sub-Antarctic notothenioid fish Eleginops maclovinus characterised by either rare or unique features. In particular, the distal site of the α chain results to be very unusual since the distal His is displaced from its canonical position. This displacement is coupled with a shortening of the highly conserved E helix and the formation of novel interactions at tertiary structure level. Interestingly, the quaternary structure is closer to the T-deoxy state of Hbs than to the R-state despite the full coordination of all chains. Notably, these peculiar structural features provide a rationale for some spectroscopic properties exhibited by the protein in solution. Finally, this unexpected structural plasticity of the heme distal side has been associated with specific sequence signatures of various Hbs.
- Institute of Biostructures and Bioimaging, CNR, Via Mezzocannone 16, Naples, Italy.
Organizational Affiliation: