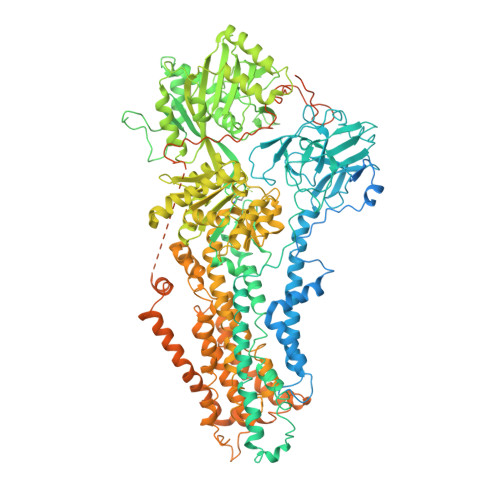
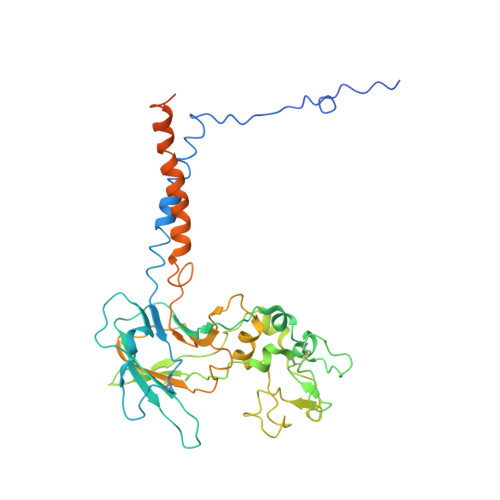
Structure and autoregulation of a P4-ATPase lipid flippase.
Timcenko, M., Lyons, J.A., Januliene, D., Ulstrup, J.J., Dieudonne, T., Montigny, C., Ash, M.R., Karlsen, J.L., Boesen, T., Kuhlbrandt, W., Lenoir, G., Moeller, A., Nissen, P.(2019) Nature 571: 366-370
- PubMed: 31243363
- DOI: https://doi.org/10.1038/s41586-019-1344-7
- Primary Citation of Related Structures:
6ROH, 6ROI, 6ROJ - PubMed Abstract:
Type 4 P-type ATPases (P4-ATPases) are lipid flippases that drive the active transport of phospholipids from exoplasmic or luminal leaflets to cytosolic leaflets of eukaryotic membranes. The molecular architecture of P4-ATPases and the mechanism through which they recognize and transport lipids have remained unknown. Here we describe the cryo-electron microscopy structure of the P4-ATPase Drs2p-Cdc50p, a Saccharomyces cerevisiae lipid flippase that is specific to phosphatidylserine and phosphatidylethanolamine. Drs2p-Cdc50p is autoinhibited by the C-terminal tail of Drs2p, and activated by the lipid phosphatidylinositol-4-phosphate (PtdIns4P or PI4P). We present three structures that represent the complex in an autoinhibited, an intermediate and a fully activated state. The analysis highlights specific features of P4-ATPases and reveals sites of autoinhibition and PI4P-dependent activation. We also observe a putative lipid translocation pathway in this flippase that involves a conserved PISL motif in transmembrane segment 4 and polar residues of transmembrane segments 2 and 5, in particular Lys1018, in the centre of the lipid bilayer.
- DANDRITE, Nordic EMBL Partnership for Molecular Medicine, Department of Molecular Biology and Genetics, Aarhus University, Aarhus, Denmark.
Organizational Affiliation: