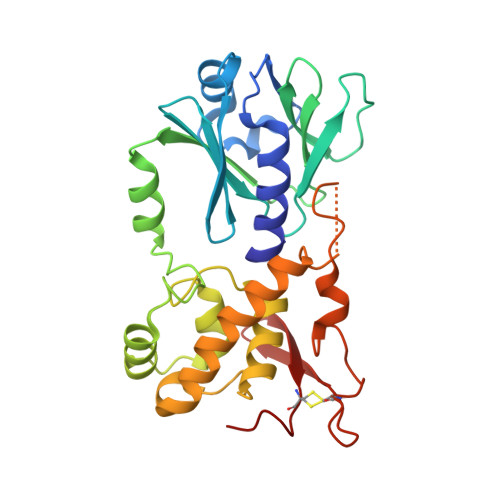
Thiopurine Derivative-Induced Fpg/Nei DNA Glycosylase Inhibition: Structural, Dynamic and Functional Insights.
Rieux, C., Goffinont, S., Coste, F., Tber, Z., Cros, J., Roy, V., Guerin, M., Gaudon, V., Bourg, S., Biela, A., Aucagne, V., Agrofoglio, L., Garnier, N., Castaing, B.(2020) Int J Mol Sci 21
- PubMed: 32192183
- DOI: https://doi.org/10.3390/ijms21062058
- Primary Citation of Related Structures:
6RNM, 6RNO, 6RNR, 6RO2, 6ROK, 6RP0, 6RP7 - PubMed Abstract:
DNA glycosylases are emerging as relevant pharmacological targets in inflammation, cancer and neurodegenerative diseases. Consequently, the search for inhibitors of these enzymes has become a very active research field. As a continuation of previous work that showed that 2-thioxanthine (2TX) is an irreversible inhibitor of zinc finger (ZnF)-containing Fpg/Nei DNA glycosylases, we designed and synthesized a mini-library of 2TX-derivatives (TXn) and evaluated their ability to inhibit Fpg/Nei enzymes. Among forty compounds, four TXn were better inhibitors than 2TX for Fpg. Unexpectedly, but very interestingly, two dithiolated derivatives more selectively and efficiently inhibit the zincless finger (ZnLF)-containing enzymes (human and mimivirus Neil1 DNA glycosylases hNeil1 and MvNei1, respectively). By combining chemistry, biochemistry, mass spectrometry, blind and flexible docking and X-ray structure analysis, we localized new TXn binding sites on Fpg/Nei enzymes. This endeavor allowed us to decipher at the atomic level the mode of action for the best TXn inhibitors on the ZnF-containing enzymes. We discovered an original inhibition mechanism for the ZnLF-containing Fpg/Nei DNA glycosylases by disulfide cyclic trimeric forms of dithiopurines. This work paves the way for the design and synthesis of a new structural class of inhibitors for selective pharmacological targeting of hNeil1 in cancer and neurodegenerative diseases.
- Centre de Biophysique Moléculaire, UPR4301 CNRS, rue Charles Sadron, CEDEX 2, F-45071 Orléans, France.
Organizational Affiliation: