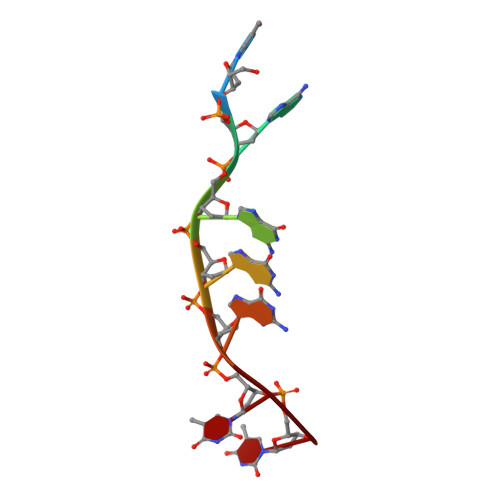
Three thymine/adenine binding modes of the ruthenium complex Lambda-[Ru(TAP)2(dppz)]2+to the G-quadruplex forming sequence d(TAGGGTT) shown by X-ray crystallography.
McQuaid, K., Hall, J.P., Baumgaertner, L., Cardin, D.J., Cardin, C.J.(2019) Chem Commun (Camb) 55: 9116-9119
- PubMed: 31298665
- DOI: https://doi.org/10.1039/c9cc04316k
- Primary Citation of Related Structures:
6RNL - PubMed Abstract:
Λ-[Ru(TAP)2(dppz)]2+ was crystallised with the G-quadruplex-forming heptamer d(TAGGGTT). Surprisingly, even though there are four unique binding sites, the complex is not in contact with any G-quartet surface. Two complexes stabilise cavities formed from terminal T·A and T·T mismatched pairs. A third shows kinking by a TAP ligand between T·T linkages, while the fourth shows sandwiching of a dppz ligand between a T·A/T·A quadruplex and a T·T mismatch, stabilised by an additional T·A base pair stacking interaction on a TAP surface. Overall, the structure shows an unexpected affinity for thymine, and suggests models for G-quadruplex loop binding.
- Department of Chemistry, University of Reading, Whiteknights, Reading, RG6 6AD, UK. c.j.cardin@reading.ac.uk.
Organizational Affiliation: