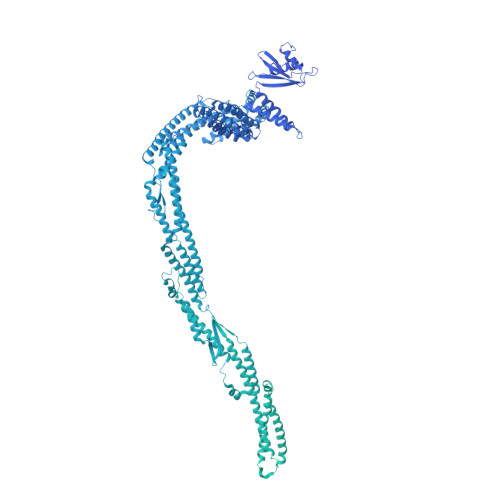
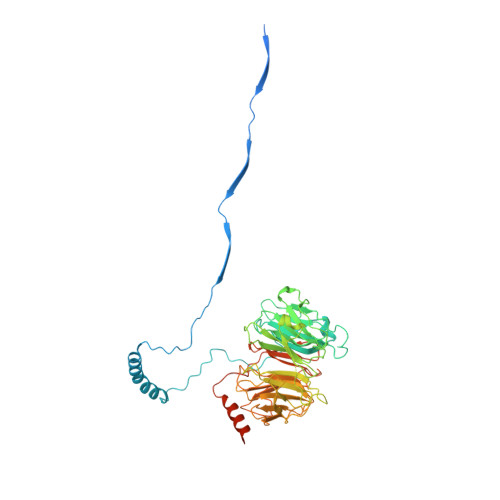
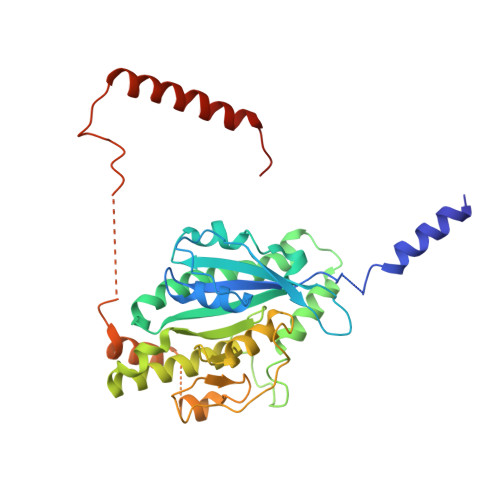
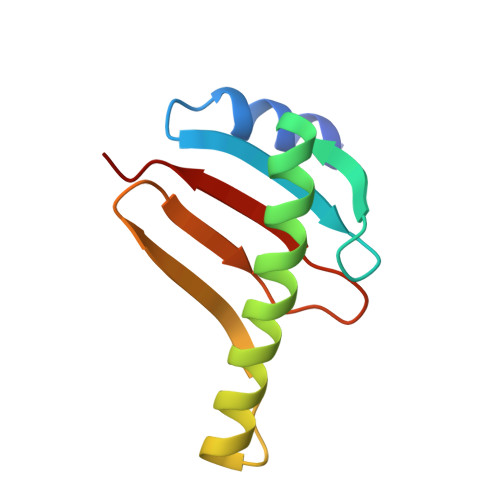
Structure of the dynein-2 complex and its assembly with intraflagellar transport trains.
Toropova, K., Zalyte, R., Mukhopadhyay, A.G., Mladenov, M., Carter, A.P., Roberts, A.J.(2019) Nat Struct Mol Biol 26: 823-829
- PubMed: 31451806
- DOI: https://doi.org/10.1038/s41594-019-0286-y
- Primary Citation of Related Structures:
6RLA, 6RLB, 6SC2 - PubMed Abstract:
Dynein-2 assembles with polymeric intraflagellar transport (IFT) trains to form a transport machinery that is crucial for cilia biogenesis and signaling. Here we recombinantly expressed the ~1.4-MDa human dynein-2 complex and solved its cryo-EM structure to near-atomic resolution. The two identical copies of the dynein-2 heavy chain are contorted into different conformations by a WDR60-WDR34 heterodimer and a block of two RB and six LC8 light chains. One heavy chain is steered into a zig-zag conformation, which matches the periodicity of the anterograde IFT-B train. Contacts between adjacent dyneins along the train indicate a cooperative mode of assembly. Removal of the WDR60-WDR34-light chain subcomplex renders dynein-2 monomeric and relieves autoinhibition of its motility. Our results converge on a model in which an unusual stoichiometry of non-motor subunits controls dynein-2 assembly, asymmetry, and activity, giving mechanistic insight into the interaction of dynein-2 with IFT trains and the origin of diverse functions in the dynein family.
- Institute of Structural and Molecular Biology, Birkbeck, University of London, London, UK.
Organizational Affiliation: