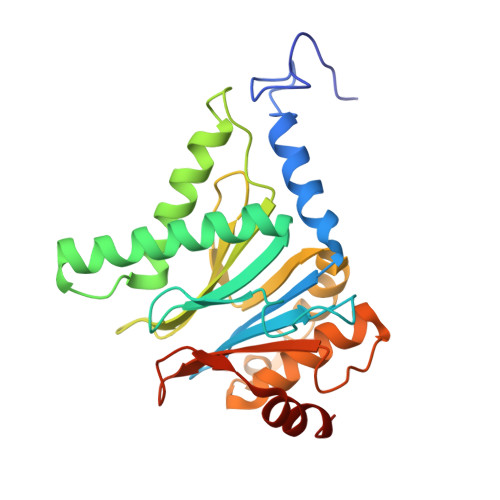
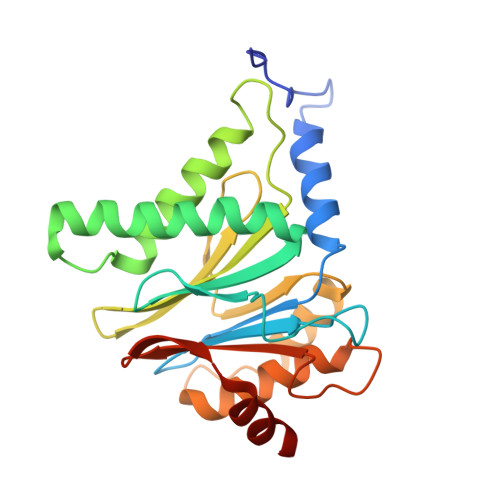
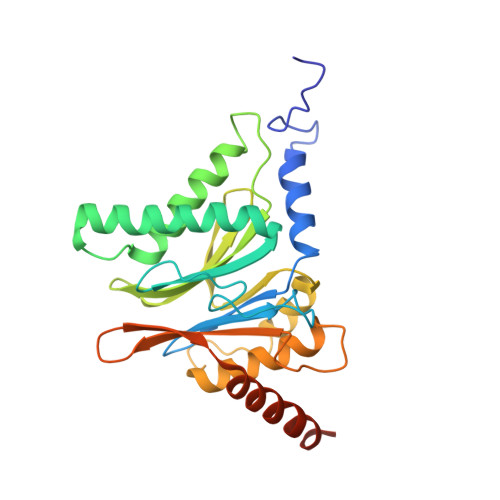
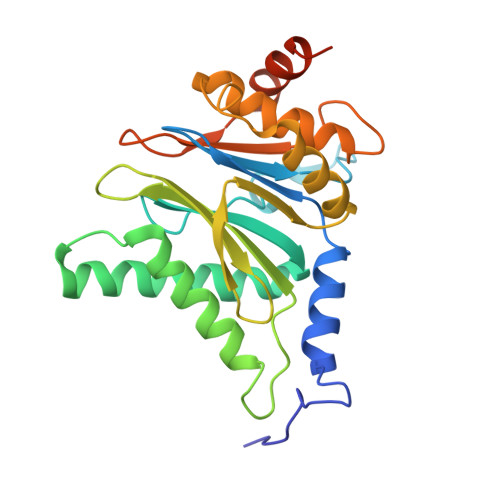
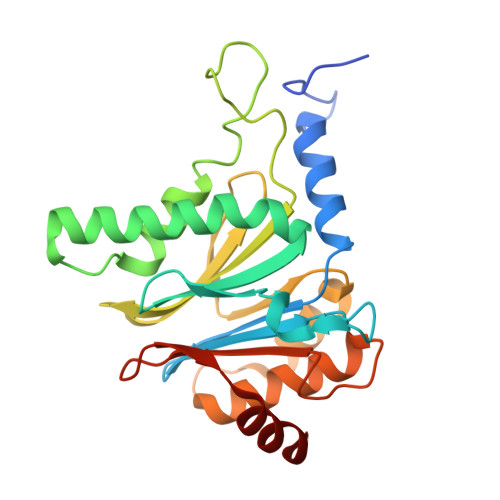
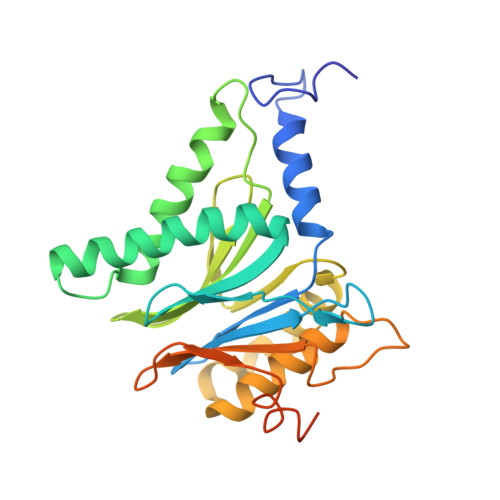
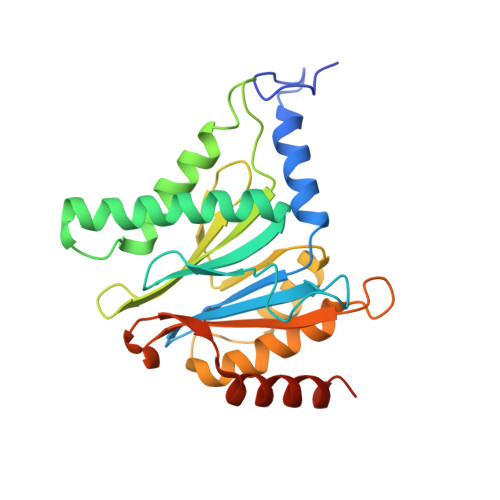
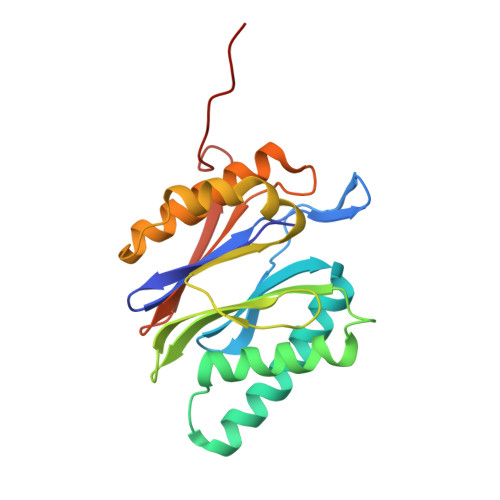
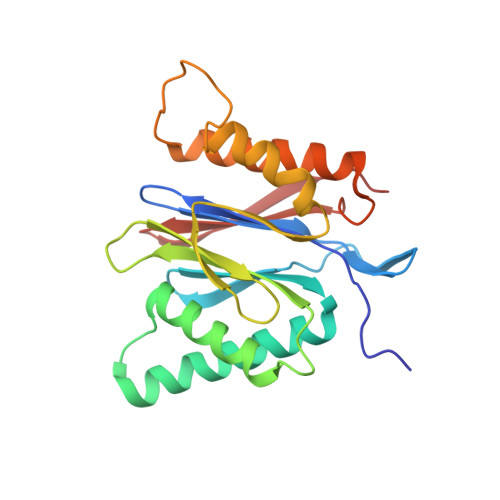
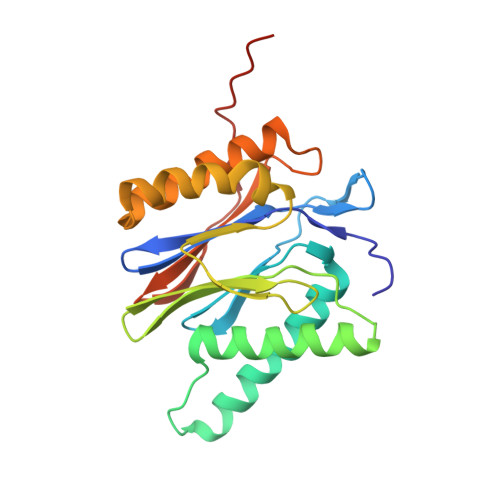
Characterization of Fully Recombinant Human 20S and 20S-PA200 Proteasome Complexes.
Toste Rego, A., da Fonseca, P.C.A.(2019) Mol Cell 76: 138-147.e5
- PubMed: 31473102
- DOI: https://doi.org/10.1016/j.molcel.2019.07.014
- Primary Citation of Related Structures:
6REY, 6RGQ - PubMed Abstract:
Proteasomes are essential in all eukaryotic cells. However, their function and regulation remain considerably elusive, particularly those of less abundant variants. We demonstrate the human 20S proteasome recombinant assembly and confirmed the recombinant complex integrity biochemically and with a 2.6 Å resolution cryo-EM map. To assess its competence to form higher-order assemblies, we prepared and analyzed recombinant human 20S-PA200, a poorly characterized nuclear complex. Its 3.0 Å resolution cryo-EM structure reveals the PA200 unique architecture; the details of its intricate interactions with the proteasome, resulting in unparalleled proteasome α ring rearrangements; and the molecular basis for PA200 allosteric modulation of the proteasome active sites. Non-protein cryo-EM densities could be assigned to PA200-bound inositol phosphates, and we speculate regarding their functional role. Here we open extensive opportunities to study the fundamental properties of the diverse and distinct eukaryotic proteasome variants and to improve proteasome targeting under different therapeutic conditions.
- MRC Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge CB2 0QH, UK.
Organizational Affiliation: