NMR resonance assignments of apo and EGRCK-inhibited factor IXa
Sendall, T.J., Huntington, J.A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Coagulation factor IX | A [auth E] | 62 | Homo sapiens | Mutation(s): 0 Gene Names: F9 EC: 3.4.21.22 | 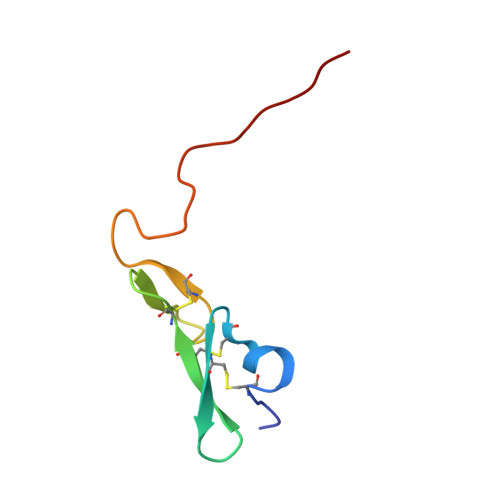 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P00740 (Homo sapiens) Explore P00740 Go to UniProtKB: P00740 | |||||
PHAROS: P00740 GTEx: ENSG00000101981 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00740 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Coagulation factor IX | B [auth S] | 235 | Homo sapiens | Mutation(s): 2 Gene Names: F9 EC: 3.4.21.22 | 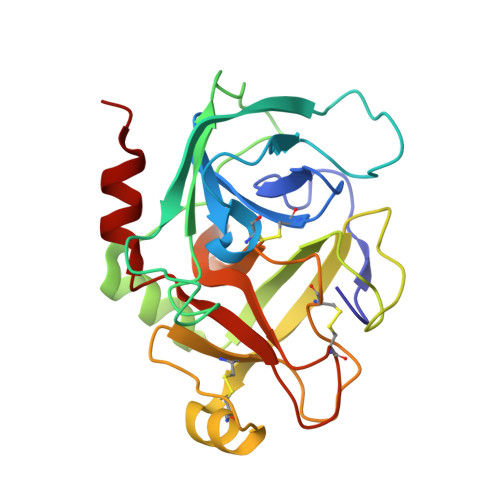 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P00740 (Homo sapiens) Explore P00740 Go to UniProtKB: P00740 | |||||
PHAROS: P00740 GTEx: ENSG00000101981 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00740 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| GLU-GLY-AR7-0QE | C [auth I] | 4 | synthetic construct | Mutation(s): 0 | 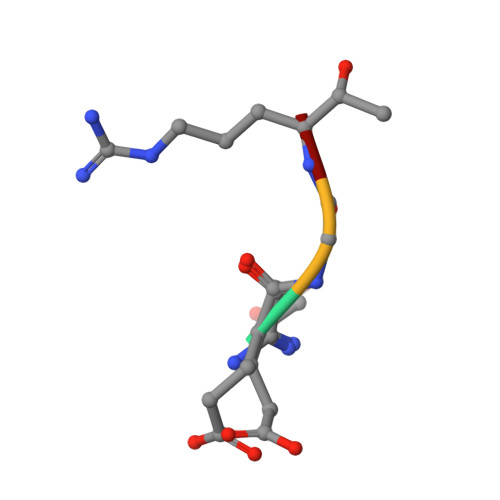 |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| B3P Query on B3P | D [auth E] | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL C11 H26 N2 O6 HHKZCCWKTZRCCL-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | F [auth S] G [auth S] H [auth S] I [auth S] J [auth S] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| CA Query on CA | E [auth S] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| AR7 Query on AR7 | C [auth I] | PEPTIDE-LIKE | C6 H17 N4 O2 |  | ARG |
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_000288 (0GJ) Query on PRD_000288 | C [auth I] | GLU-GLY-ARG-CHLOROMETHYL KETONE | Peptide-like / Inhibitor |  | |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 100.16 | α = 90 |
| b = 100.16 | β = 90 |
| c = 91.5 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| MOSFLM | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| British Heart Foundation | United Kingdom | PG/11/91/29117 |