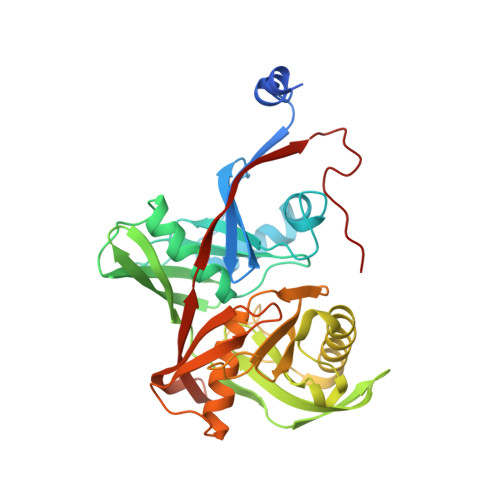
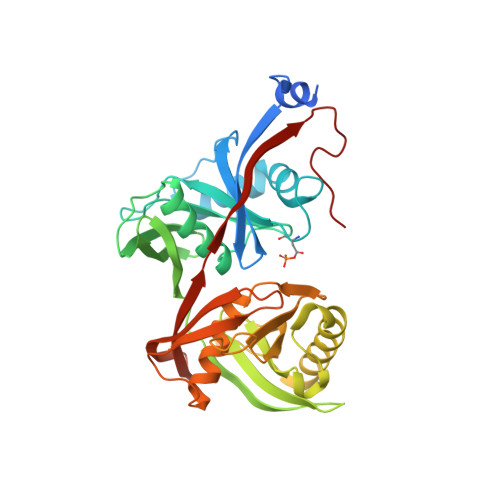
Structure of Thermococcus litoralis trans-3-hydroxy-l-proline dehydratase in the free and substrate-complexed form.
Ferraris, D.M., Miggiano, R., Watanabe, S., Rizzi, M.(2019) Biochem Biophys Res Commun 516: 189-195
- PubMed: 31208721
- DOI: https://doi.org/10.1016/j.bbrc.2019.06.021
- Primary Citation of Related Structures:
6R76, 6R77 - PubMed Abstract:
Hydroxyprolines (Hyp) are non-standard amino acids derived from the post-translational modification of proteins by prolyl hydroxylase enzymes. Some plants and bacteria produce Hyp, and the isomers trans-3-Hydroxy-l-proline (T3LHyp) and trans-4-Hydroxy-l-proline (T4LHyp) are major components of mammalian collagen. While T4LHyp is metabolised following distinct degradative pathways in mammals and bacteria, T3LHyp metabolic pathway is conserved in bacteria, plants and mammals, and involves a T3LHyp dehydratase (T3LHypD) in the first degradation step. We report here the crystal structure of T3LHypD from the archaea Thermococcus litoralis in the free and substrate-complexed form. The model shows an "open" and a "closed" conformation depending on the presence (or absence) of the substrate in the catalytic site and allows the mapping of the residues involved in ligand recognition. Moreover, the structure highlights the presence of a water molecule interacting with the hydroxy group of the substrate and potentially involved in catalysis. The structure here reported is the first of its family to be elucidated, and represents a valid model for rationalising the substrate specificity and catalysis of T3LHyp dehydratases.
- Dipartimento di Scienze del Farmaco, Università del Piemonte Orientale, Via Bovio 6, 28100, Novara, Italy; IXTAL srl, Via Bovio 6, 28100, Novara, Italy. Electronic address: davide.ferraris@uniupo.it.
Organizational Affiliation: