Targeting farnesyl pyrophosphate synthase of Trypanosoma cruzi by fragment-based lead discovery
Petrick, J.K., Jahnke, W.(2019) Thesis
Experimental Data Snapshot
Starting Model: experimental
View more details
(2019) Thesis
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Farnesyl pyrophosphate synthase | 369 | Trypanosoma brucei | Mutation(s): 0 | 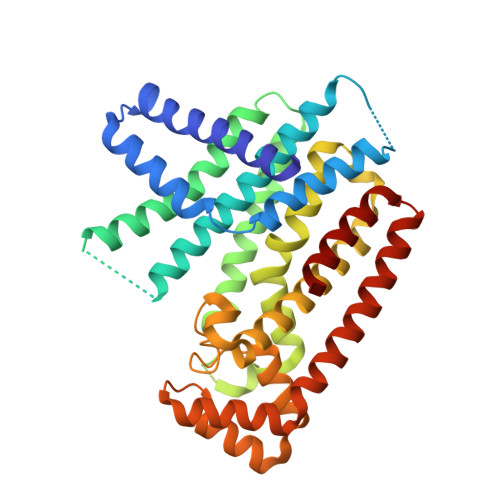 | |
UniProt | |||||
Find proteins for Q86C09 (Trypanosoma brucei) Explore Q86C09 Go to UniProtKB: Q86C09 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q86C09 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| BFH Query on BFH | B [auth A] | 1-(carboxymethyl)-1H-benzo[g]indole-2-carboxylic acid C15 H11 N O4 AKGLRHRNGGDMDM-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 61.336 | α = 90 |
| b = 61.336 | β = 90 |
| c = 340.077 | γ = 120 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| BUSTER | refinement |
| PDB_EXTRACT | data extraction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Marie Sklodowska-Curie Actions, FragNET ITN | Switzerland | 675899 |