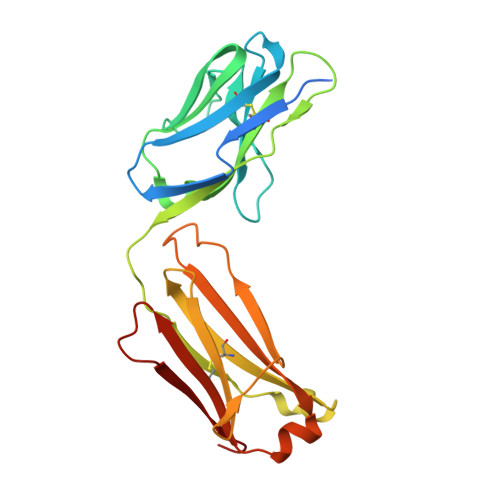
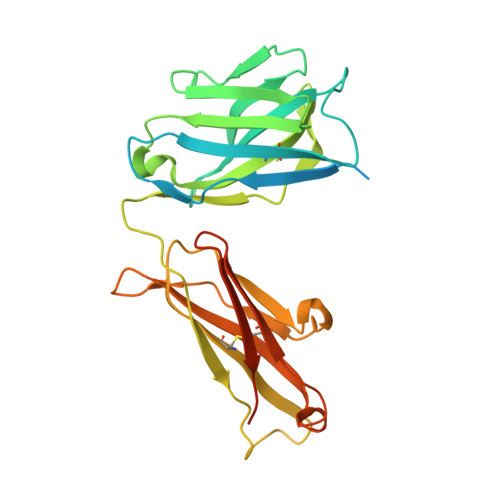
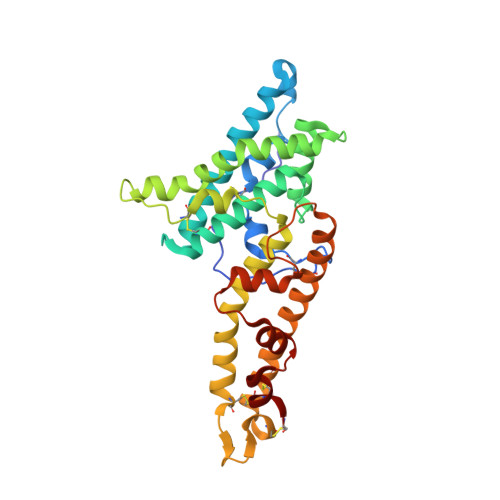
Structural basis for inhibition of Plasmodium vivax invasion by a broadly neutralizing vaccine-induced human antibody.
Rawlinson, T.A., Barber, N.M., Mohring, F., Cho, J.S., Kosaisavee, V., Gerard, S.F., Alanine, D.G.W., Labbe, G.M., Elias, S.C., Silk, S.E., Quinkert, D., Jin, J., Marshall, J.M., Payne, R.O., Minassian, A.M., Russell, B., Renia, L., Nosten, F.H., Moon, R.W., Higgins, M.K., Draper, S.J.(2019) Nat Microbiol 4: 1497-1507
- PubMed: 31133755
- DOI: https://doi.org/10.1038/s41564-019-0462-1
- Primary Citation of Related Structures:
6R2S - PubMed Abstract:
The most widespread form of malaria is caused by Plasmodium vivax. To replicate, this parasite must invade immature red blood cells through a process requiring interaction of the P. vivax Duffy binding protein (PvDBP) with its human receptor, the Duffy antigen receptor for chemokines. Naturally acquired antibodies that inhibit this interaction associate with clinical immunity, suggesting PvDBP as a leading candidate for inclusion in a vaccine to prevent malaria due to P. vivax. Here, we isolated a panel of monoclonal antibodies from human volunteers immunized in a clinical vaccine trial of PvDBP. We screened their ability to prevent PvDBP from binding to the Duffy antigen receptor for chemokines, and their capacity to block red blood cell invasion by a transgenic Plasmodium knowlesi parasite genetically modified to express PvDBP and to prevent reticulocyte invasion by multiple clinical isolates of P. vivax. This identified a broadly neutralizing human monoclonal antibody that inhibited invasion of all tested strains of P. vivax. Finally, we determined the structure of a complex of this antibody bound to PvDBP, indicating the molecular basis for inhibition. These findings will guide future vaccine design strategies and open up possibilities for testing the prophylactic use of such an antibody.
- Jenner Institute, University of Oxford, Oxford, UK.
Organizational Affiliation: