Atomic description of an enzyme reaction dependent on reversible 1,3-dipolar cycloaddition
Bailey, S.S., Payne, K.A.P., Saaret, A., Marshall, S.A., Gostimskaya, I., Kosov, I., Fisher, K., Hay, S., Leys, D.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ferulic acid decarboxylase 1 | 508 | Aspergillus niger | Mutation(s): 0 Gene Names: fdc1, An03g06590 EC: 4.1.1.102 | 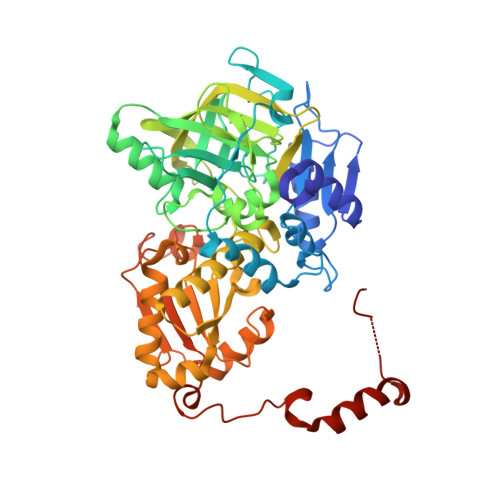 | |
UniProt | |||||
Find proteins for A2QHE5 (Aspergillus niger (strain ATCC MYA-4892 / CBS 513.88 / FGSC A1513)) Explore A2QHE5 Go to UniProtKB: A2QHE5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A2QHE5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| BYN (Subject of Investigation/LOI) Query on BYN | E [auth A] | hydroxylated prenyl-FMN C22 H31 N4 O10 P BDNVCRCOZIAGID-LWGWVAHUSA-N |  | ||
| 4LW (Subject of Investigation/LOI) Query on 4LW | F [auth A] | (2Z)-2-fluoro-3-phenylprop-2-enoic acid C9 H7 F O2 QONCEXMULRJPPY-VURMDHGXSA-N |  | ||
| MN Query on MN | B [auth A] | MANGANESE (II) ION Mn WAEMQWOKJMHJLA-UHFFFAOYSA-N |  | ||
| K Query on K | C [auth A], D [auth A] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 95.681 | α = 90 |
| b = 63.945 | β = 90 |
| c = 87.573 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| xia2 | data reduction |
| xia2 | data scaling |
| Funding Organization | Location | Grant Number |
|---|---|---|
| European Research Council | United Kingdom | preFAB 695013 |
| Biotechnology and Biological Sciences Research Council | United Kingdom | BB/K017802 |