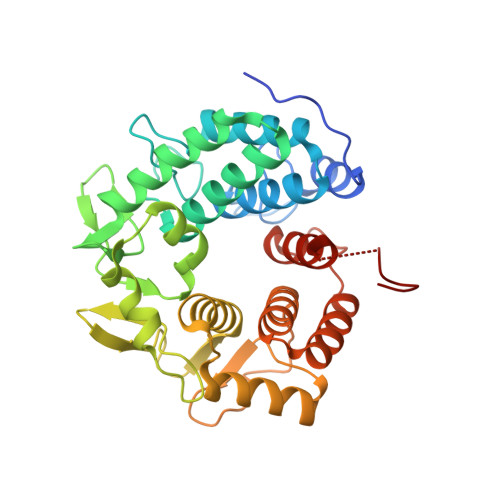
Crystal structure of the glycoside hydrolase PssZ from Listeria monocytogenes.
Wu, H., Qiao, S., Li, D., Guo, L., Zhu, M., Ma, L.Z.(2019) Acta Crystallogr F Struct Biol Commun 75: 501-506
- PubMed: 31282870
- DOI: https://doi.org/10.1107/S2053230X19008100
- Primary Citation of Related Structures:
6R2M - PubMed Abstract:
Biofilms are microbial communities that are embedded in the extracellular matrix. The exopolysaccharide (EPS) is a key component of the biofilm matrix that maintains the structure of the biofilm and protects the bacteria from antimicrobials. Microbial glycoside hydrolases have been exploited to disrupt biofilms by breaking down EPSs. PssZ has recently been identified as a glycoside hydrolase that can disperse aggregates of Listeria monocytogenes. In this study, the crystal structure of PssZ has been determined at 1.6 Å resolution. PssZ belongs to glycoside hydrolase family 8 and adopts a classical (α/α) 6 -barrel fold. This architecture forms a deep groove which may serve as the substrate-binding pocket. The conserved catalytic residues (Glu72, Trp110, Asn119, Phe167, Tyr183 and Asp232) are localized at the centre of the groove. This crystal structure will help to improve the understanding of the hydrolytic mechanism of PssZ and its application as a biofilm disrupter.
- State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, People's Republic of China.
Organizational Affiliation: