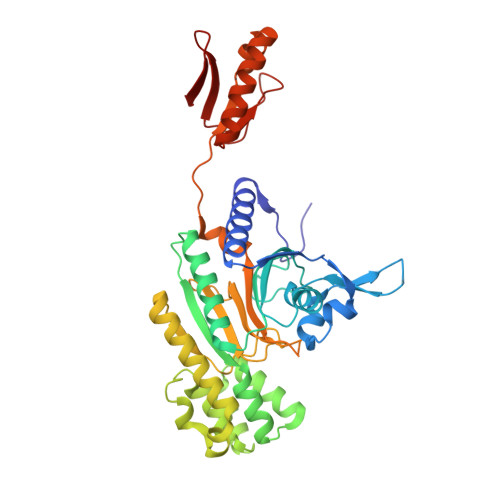
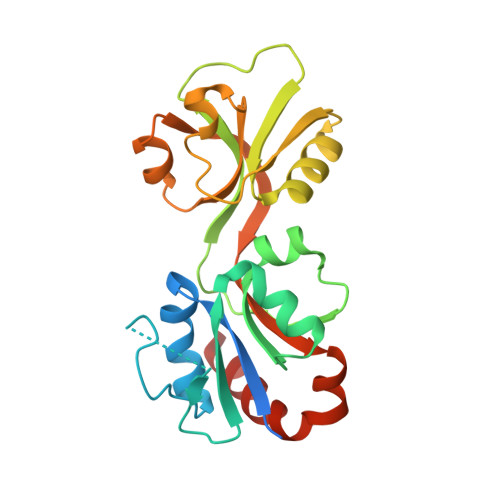
Mapping the Structural Path for Allosteric Inhibition of a Short-Form ATP Phosphoribosyltransferase by Histidine.
Thomson, C.M., Alphey, M.S., Fisher, G., da Silva, R.G.(2019) Biochemistry 58: 3078-3086
- PubMed: 31251578
- DOI: https://doi.org/10.1021/acs.biochem.9b00282
- Primary Citation of Related Structures:
6R02 - PubMed Abstract:
ATP phosphoribosyltransferase (ATPPRT) catalyzes the first step of histidine biosynthesis, being allosterically inhibited by the final product of the pathway. Allosteric inhibition of long-form ATPPRTs by histidine has been extensively studied, but inhibition of short-form ATPPRTs is poorly understood. Short-form ATPPRTs are hetero-octamers formed by four catalytic subunits (HisG S ) and four regulatory subunits (HisZ). HisG S alone is catalytically active and insensitive to histidine. HisZ enhances catalysis by HisG S in the absence of histidine but mediates allosteric inhibition in its presence. Here, steady-state and pre-steady-state kinetics establish that histidine is a noncompetitive inhibitor of short-form ATPPRT and that inhibition does not occur by dissociating HisG S from the hetero-octamer. The crystal structure of ATPPRT in complex with histidine and the substrate 5-phospho-α-d-ribosyl-1-pyrophosphate was determined, showing histidine bound solely to HisZ, with four histidine molecules per hetero-octamer. Histidine binding involves the repositioning of two HisZ loops. The histidine-binding loop moves closer to histidine to establish polar contacts. This leads to a hydrogen bond between its Tyr263 and His104 in the Asp101-Leu117 loop. The Asp101-Leu117 loop leads to the HisZ-HisG S interface, and in the absence of histidine, its motion prompts HisG S conformational changes responsible for catalytic activation. Following histidine binding, interaction with the histidine-binding loop may prevent the Asp101-Leu117 loop from efficiently sampling conformations conducive to catalytic activation. Tyr263Phe- Pa HisZ-containing Pa ATPPRT, however, is less susceptible though not insensitive to histidine inhibition, suggesting the Tyr263-His104 interaction may be relevant to yet not solely responsible for transmission of the allosteric signal.
- School of Biology, Biomedical Sciences Research Complex , University of St Andrews , St Andrews , Fife KY16 9ST , U.K.
Organizational Affiliation: