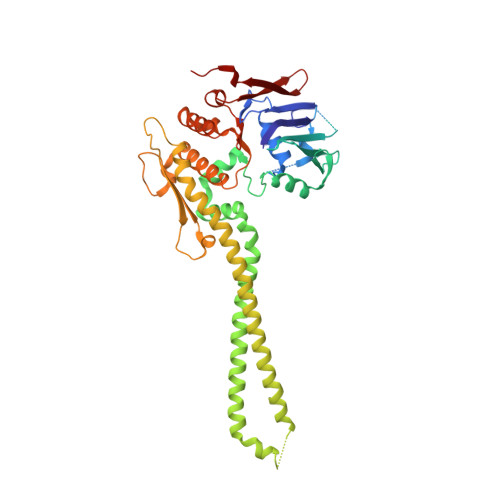
The structure of the cohesin ATPase elucidates the mechanism of SMC-kleisin ring opening.
Muir, K.W., Li, Y., Weis, F., Panne, D.(2020) Nat Struct Mol Biol 27: 233-239
- PubMed: 32066964
- DOI: https://doi.org/10.1038/s41594-020-0379-7
- Primary Citation of Related Structures:
6QPQ, 6QPW - PubMed Abstract:
Genome regulation requires control of chromosome organization by SMC-kleisin complexes. The cohesin complex contains the Smc1 and Smc3 subunits that associate with the kleisin Scc1 to form a ring-shaped complex that can topologically engage chromatin to regulate chromatin structure. Release from chromatin involves opening of the ring at the Smc3-Scc1 interface in a reaction that is controlled by acetylation and engagement of the Smc ATPase head domains. To understand the underlying molecular mechanisms, we have determined the 3.2-Å resolution cryo-electron microscopy structure of the ATPγS-bound, heterotrimeric cohesin ATPase head module and the 2.1-Å resolution crystal structure of a nucleotide-free Smc1-Scc1 subcomplex from Saccharomyces cerevisiae and Chaetomium thermophilium. We found that ATP-binding and Smc1-Smc3 heterodimerization promote conformational changes within the ATPase that are transmitted to the Smc coiled-coil domains. Remodeling of the coiled-coil domain of Smc3 abrogates the binding surface for Scc1, thus leading to ring opening at the Smc3-Scc1 interface.
- European Molecular Biology Laboratory, Grenoble, France. kmuir@mrc-lmb.cam.ac.uk.
Organizational Affiliation: