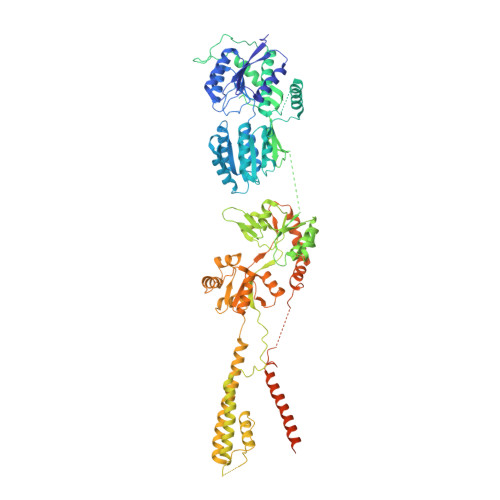
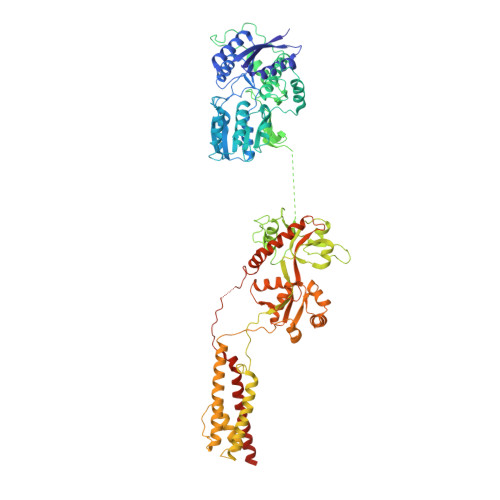
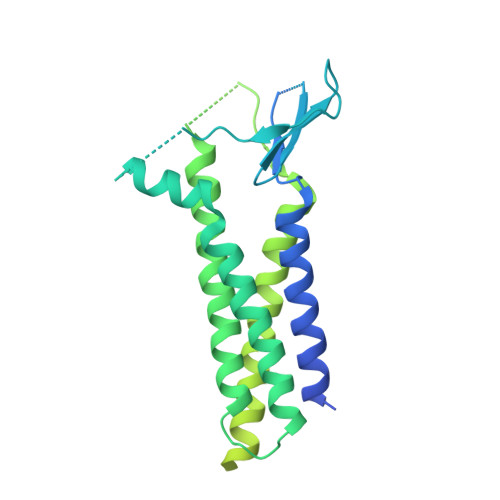
Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP gamma 8.
Herguedas, B., Watson, J.F., Ho, H., Cais, O., Garcia-Nafria, J., Greger, I.H.(2019) Science 364
- PubMed: 30872532
- DOI: https://doi.org/10.1126/science.aav9011
- Primary Citation of Related Structures:
6QKC, 6QKZ - PubMed Abstract:
AMPA-type glutamate receptors (AMPARs) mediate excitatory neurotransmission and are central regulators of synaptic plasticity, a molecular mechanism underlying learning and memory. Although AMPARs act predominantly as heteromers, structural studies have focused on homomeric assemblies. Here, we present a cryo-electron microscopy structure of the heteromeric GluA1/2 receptor associated with two transmembrane AMPAR regulatory protein (TARP) γ8 auxiliary subunits, the principal AMPAR complex at hippocampal synapses. Within the receptor, the core subunits arrange to give the GluA2 subunit dominant control of gating. This structure reveals the geometry of the Q/R site that controls calcium flux, suggests association of TARP-stabilized lipids, and demonstrates that the extracellular loop of γ8 modulates gating by selectively interacting with the GluA2 ligand-binding domain. Collectively, this structure provides a blueprint for deciphering the signal transduction mechanisms of synaptic AMPARs.
- Neurobiology Division, Medical Research Council (MRC) Laboratory of Molecular Biology, Cambridge, UK. ig@mrc-lmb.cam.ac.uk hergueda@mrc-lmb.cam.ac.uk.
Organizational Affiliation: