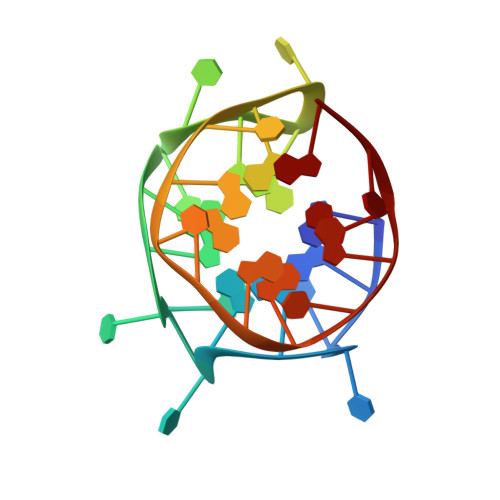
NMR solution and X-ray crystal structures of a DNA molecule containing both right- and left-handed parallel-stranded G-quadruplexes.
Winnerdy, F.R., Bakalar, B., Maity, A., Vandana, J.J., Mechulam, Y., Schmitt, E., Phan, A.T.(2019) Nucleic Acids Res 47: 8272-8281
- PubMed: 31216034
- DOI: https://doi.org/10.1093/nar/gkz349
- Primary Citation of Related Structures:
6JCE, 6QJO - PubMed Abstract:
Analogous to the B- and Z-DNA structures in double-helix DNA, there exist both right- and left-handed quadruple-helix (G-quadruplex) DNA. Numerous conformations of right-handed and a few left-handed G-quadruplexes were previously observed, yet they were always identified separately. Here, we present the NMR solution and X-ray crystal structures of a right- and left-handed hybrid G-quadruplex. The structure reveals a stacking interaction between two G-quadruplex blocks with different helical orientations and displays features of both right- and left-handed G-quadruplexes. An analysis of loop mutations suggests that single-nucleotide loops are preferred or even required for the left-handed G-quadruplex formation. The discovery of a right- and left-handed hybrid G-quadruplex further expands the polymorphism of G-quadruplexes and is potentially useful in designing a left-to-right junction in G-quadruplex engineering.
- School of Physical and Mathematical Sciences, Nanyang Technological University, Singapore 637371, Singapore.
Organizational Affiliation: