Structural evidence for an essential Fe-S cluster in the catalytic core domain of DNA polymerase ε.
Ter Beek, J., Parkash, V., Bylund, G.O., Osterman, P., Sauer-Eriksson, A.E., Johansson, E.(2019) Nucleic Acids Res 47: 5712-5722
- PubMed: 30968138
- DOI: https://doi.org/10.1093/nar/gkz248
- Primary Citation of Related Structures:
6H1V, 6QIB - PubMed Abstract:
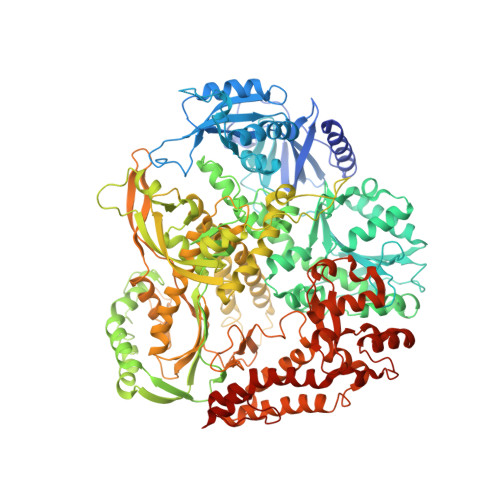
DNA polymerase ϵ (Pol ϵ), the major leading-strand DNA polymerase in eukaryotes, has a catalytic subunit (Pol2) and three non-catalytic subunits. The N-terminal half of Pol2 (Pol2CORE) exhibits both polymerase and exonuclease activity. It has been suggested that both the non-catalytic C-terminal domain of Pol2 (with the two cysteine motifs CysA and CysB) and Pol2CORE (with the CysX cysteine motif) are likely to coordinate an Fe-S cluster. Here, we present two new crystal structures of Pol2CORE with an Fe-S cluster bound to the CysX motif, supported by an anomalous signal at that position. Furthermore we show that purified four-subunit Pol ϵ, Pol ϵ CysAMUT (C2111S/C2133S), and Pol ϵ CysBMUT (C2167S/C2181S) all have an Fe-S cluster that is not present in Pol ϵ CysXMUT (C665S/C668S). Pol ϵ CysAMUT and Pol ϵ CysBMUT behave similarly to wild-type Pol ϵ in in vitro assays, but Pol ϵ CysXMUT has severely compromised DNA polymerase activity that is not the result of an excessive exonuclease activity. Tetrad analyses show that haploid yeast strains carrying CysXMUT are inviable. In conclusion, Pol ϵ has a single Fe-S cluster bound at the base of the P-domain, and this Fe-S cluster is essential for cell viability and polymerase activity.
- Department of Medical Biochemistry and Biophysics, Umeå University, Umeå 90187, Sweden.
Organizational Affiliation: