Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Wrobel, A.G., Kadlecova, Z., Kamenicky, J., Yang, J.C., Herrmann, T., Kelly, B.T., McCoy, A.J., Evans, P.R., Martin, S., Muller, S., Sroubek, F., Neuhaus, D., Honing, S., Owen, D.J.(2019) Dev Cell 50: 494-508.e11
- PubMed: 31430451
- DOI: https://doi.org/10.1016/j.devcel.2019.07.017
- Primary Citation of Related Structures:
6QH5, 6QH6, 6QH7, 6RH5, 6RH6 - PubMed Abstract:
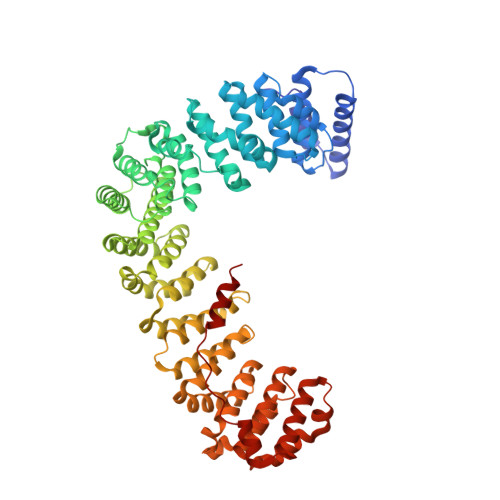
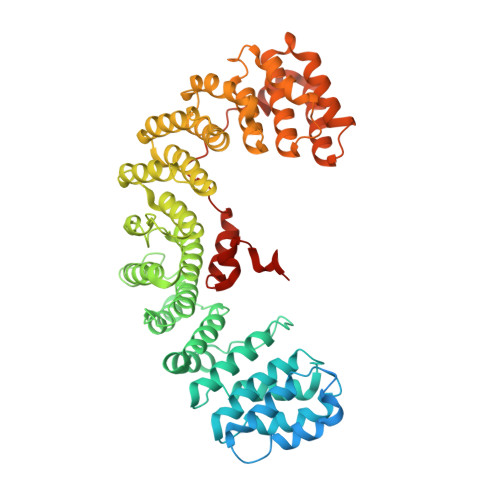
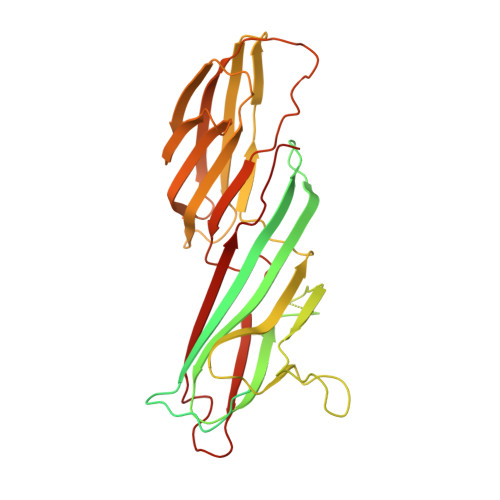
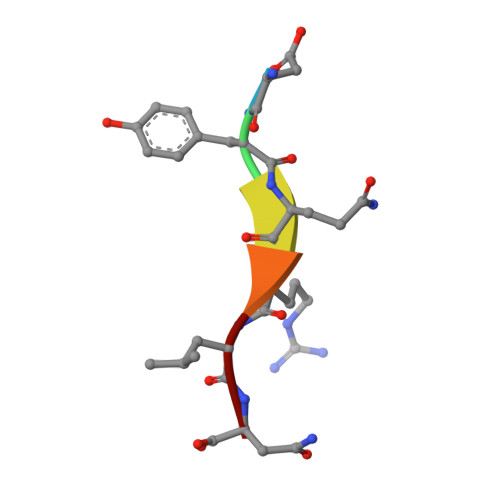
Clathrin-mediated endocytosis (CME) is key to maintaining the transmembrane protein composition of cells' limiting membranes. During mammalian CME, a reversible phosphorylation event occurs on Thr156 of the μ2 subunit of the main endocytic clathrin adaptor, AP2. We show that this phosphorylation event starts during clathrin-coated pit (CCP) initiation and increases throughout CCP lifetime. μ2Thr156 phosphorylation favors a new, cargo-bound conformation of AP2 and simultaneously creates a binding platform for the endocytic NECAP proteins but without significantly altering AP2's cargo affinity in vitro. We describe the structural bases of both. NECAP arrival at CCPs parallels that of clathrin and increases with μ2Thr156 phosphorylation. In turn, NECAP recruits drivers of late stages of CCP formation, including SNX9, via a site distinct from where NECAP binds AP2. Disruption of the different modules of this phosphorylation-based temporal regulatory system results in CCP maturation being delayed and/or stalled, hence impairing global rates of CME.
- CIMR, WT/MRC Building, Hills Road, Cambridge CB2 0QQ, UK.
Organizational Affiliation: