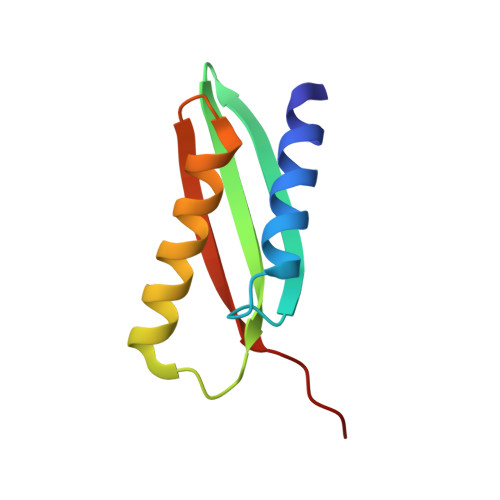
From conservation to structure, studies of magnetosome associated cation diffusion facilitators (CDF) proteins in Proteobacteria.
Keren-Khadmy, N., Zeytuni, N., Kutnowski, N., Perriere, G., Monteil, C., Zarivach, R.(2020) PLoS One 15: e0231839-e0231839
- PubMed: 32310978
- DOI: https://doi.org/10.1371/journal.pone.0231839
- Primary Citation of Related Structures:
6QFJ - PubMed Abstract:
Magnetotactic bacteria (MTB) are prokaryotes that sense the geomagnetic field lines to geolocate and navigate in aquatic sediments. They are polyphyletically distributed in several bacterial divisions but are mainly represented in the Proteobacteria. In this phylum, magnetotactic Deltaproteobacteria represent the most ancestral class of MTB. Like all MTB, they synthesize membrane-enclosed magnetic nanoparticles, called magnetosomes, for magnetic sensing. Magnetosome biogenesis is a complex process involving a specific set of genes that are conserved across MTB. Two of the most conserved genes are mamB and mamM, that encode for the magnetosome-associated proteins and are homologous to the cation diffusion facilitator (CDF) protein family. In magnetotactic Alphaproteobacteria MTB species, MamB and MamM proteins have been well characterized and play a central role in iron-transport required for biomineralization. However, their structural conservation and their role in more ancestral groups of MTB like the Deltaproteobacteria have not been established. Here we studied magnetite cluster MamB and MamM cytosolic C-terminal domain (CTD) structures from a phylogenetically distant magnetotactic Deltaproteobacteria species represented by BW-1 strain, which has the unique ability to biomineralize magnetite and greigite. We characterized them in solution, analyzed their crystal structures and compared them to those characterized in Alphaproteobacteria MTB species. We showed that despite the high phylogenetic distance, MamBBW-1 and MamMBW-1 CTDs share high structural similarity with known CDF-CTDs and will probably share a common function with the Alphaproteobacteria MamB and MamM.
- Department of Life Sciences, Ben-Gurion University of the Negev, Beer-Sheva, Israel.
Organizational Affiliation: