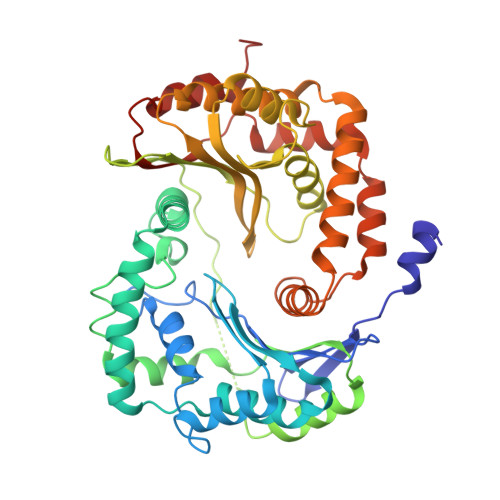
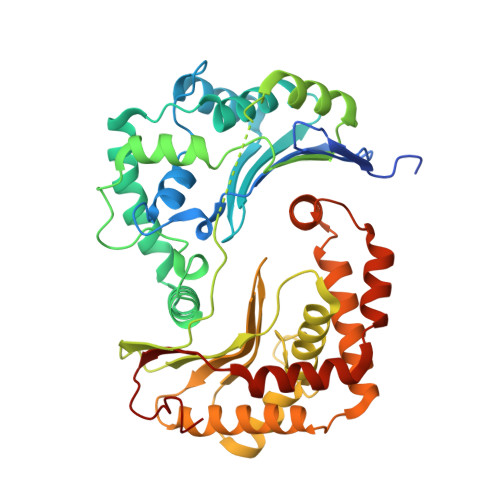
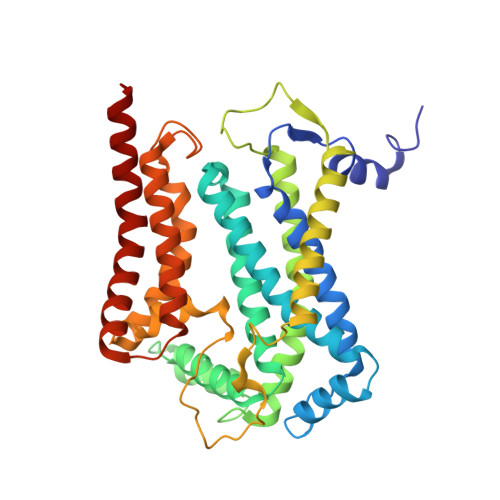
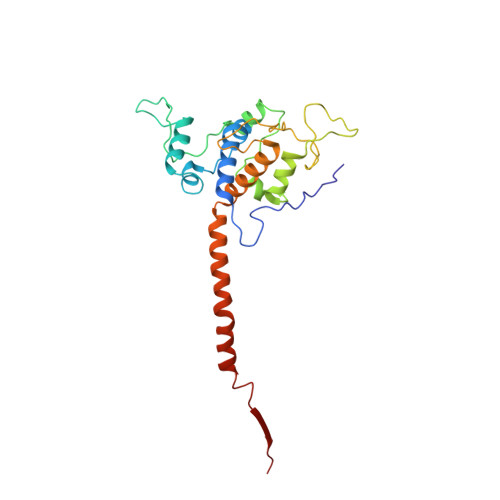
Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Letts, J.A., Fiedorczuk, K., Degliesposti, G., Skehel, M., Sazanov, L.A.(2019) Mol Cell 75: 1131-1146.e6
- PubMed: 31492636
- DOI: https://doi.org/10.1016/j.molcel.2019.07.022
- Primary Citation of Related Structures:
6Q9B, 6Q9D, 6Q9E, 6QA9, 6QBX, 6QC2, 6QC3, 6QC4, 6QC5, 6QC6, 6QC7, 6QC8, 6QC9, 6QCA, 6QCF - PubMed Abstract:
The mitochondrial electron transport chain complexes are organized into supercomplexes (SCs) of defined stoichiometry, which have been proposed to regulate electron flux via substrate channeling. We demonstrate that CoQ trapping in the isolated SC I+III 2 limits complex (C)I turnover, arguing against channeling. The SC structure, resolved at up to 3.8 Å in four distinct states, suggests that CoQ oxidation may be rate limiting because of unequal access of CoQ to the active sites of CIII 2 . CI shows a transition between "closed" and "open" conformations, accompanied by the striking rotation of a key transmembrane helix. Furthermore, the state of CI affects the conformational flexibility within CIII 2 , demonstrating crosstalk between the enzymes. CoQ was identified at only three of the four binding sites in CIII 2 , suggesting that interaction with CI disrupts CIII 2 symmetry in a functionally relevant manner. Together, these observations indicate a more nuanced functional role for the SCs.
- Institute of Science and Technology Austria, Klosterneuberg 3400, Austria; Department of Molecular and Cellular Biology, University of California, Davis, Davis, CA 95616, USA.
Organizational Affiliation: