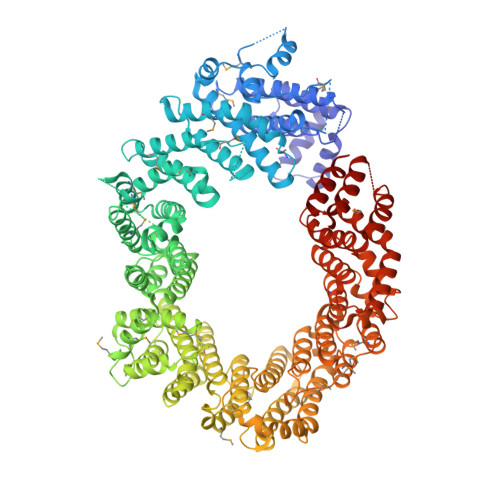
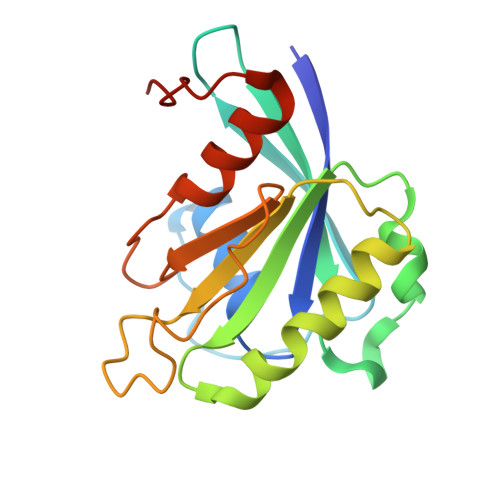
Structural basis for the nuclear import and export functions of the biportin Pdr6/Kap122.
Aksu, M., Trakhanov, S., Vera Rodriguez, A., Gorlich, D.(2019) J Cell Biol 218: 1839-1852
- PubMed: 31023722
- DOI: https://doi.org/10.1083/jcb.201812093
- Primary Citation of Related Structures:
6Q82, 6Q83, 6Q84 - PubMed Abstract:
Importins ferry proteins into nuclei while exportins carry cargoes to the cytoplasm. In the accompanying paper in this issue (Vera Rodriguez et al. 2019. J. Cell Biol. https://doi.org/10.1083/jcb.201812091), we discovered that Pdr6 is a biportin that imports, e.g., the SUMO E2 ligase Ubc9 while depleting the translation factor eIF5A from the nuclear compartment. In this paper, we report the structures of key transport intermediates, namely, of the Ubc9•Pdr6 import complex, of the RanGTP•Pdr6 heterodimer, and of the trimeric RanGTP•Pdr6•eIF5A export complex. These revealed nonlinear transport signals, chaperone-like interactions, and how the RanGTPase system drives Pdr6 to transport Ubc9 and eIF5A in opposite directions. The structures also provide unexpected insights into the evolution of transport selectivity. Specifically, they show that recognition of Ubc9 by Pdr6 differs fundamentally from that of the human Ubc9-importer Importin 13. Likewise, Pdr6 recognizes eIF5A in a nonhomologous manner compared with the mammalian eIF5A-exporter Exportin 4. This suggests that the import of Ubc9 and active nuclear exclusion of eIF5A evolved in different eukaryotic lineages more than once and independently from each other.
- Department of Cellular Logistics, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany.
Organizational Affiliation: