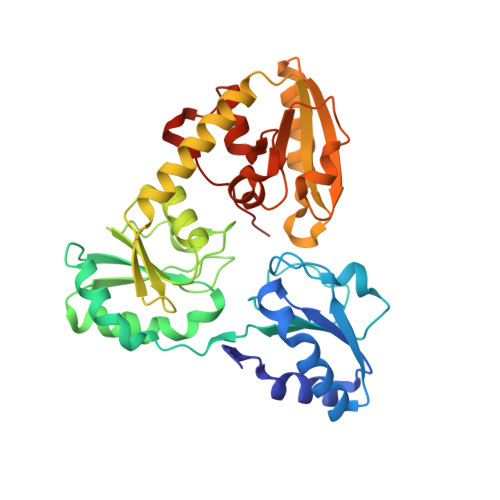
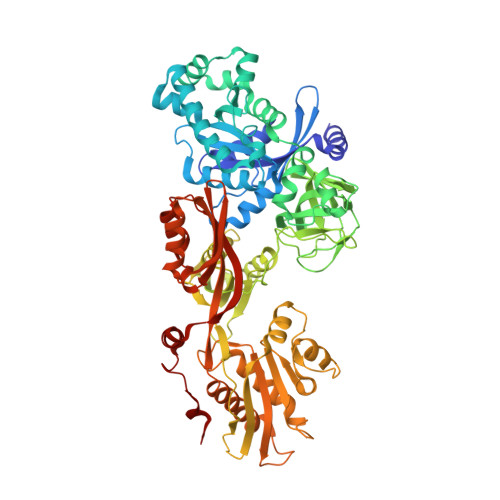
The Crystal Structure of Dph2 in Complex with Elongation Factor 2 Reveals the Structural Basis for the First Step of Diphthamide Biosynthesis.
Fenwick, M.K., Dong, M., Lin, H., Ealick, S.E.(2019) Biochemistry 58: 4343-4351
- PubMed: 31566354
- DOI: https://doi.org/10.1021/acs.biochem.9b00718
- Primary Citation of Related Structures:
6Q2D, 6Q2E - PubMed Abstract:
Elongation factor 2 (EF-2), a five-domain, GTP-dependent ribosomal translocase of archaebacteria and eukaryotes, undergoes post-translational modification to form diphthamide on a specific histidine residue in domain IV prior to binding the ribosome. The first step of diphthamide biosynthesis in archaebacteria is catalyzed by Dph2, a homodimeric radical S -adenosylmethionine (SAM) enzyme having a noncanonical architecture. Here, we describe a 3.5 Å resolution crystal structure of the Methanobrevibacter smithii ( Ms ) Dph2 homodimer bound to two molecules of Ms EF-2, one of which is ordered and the other largely disordered. Ms EF-2 is bound to both protomers of Ms Dph2, with domain IV bound to the active site of one protomer and domain III bound to a surface α-helix of an adjacent protomer. The histidine substrate of domain IV is inserted into the active site, which reveals for the first time the architecture of the Dph2 active site in complex with its target substrate. We also determined a high-resolution crystal structure of isolated Ms Dph2 bound to 5'-methylthioadenosine that shows a conserved arginine residue preoriented by conserved phenylalanine and aspartate residues for binding the carboxylate group of SAM. Mutagenesis experiments suggest that the arginine plays an important role in the first step of diphthamide biosynthesis.